Gene
KWMTBOMO13301 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004694
Annotation
vacuolar_protein_sorting-associated_protein_VTA1_homolog_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.271 Nuclear Reliability : 1.705
Sequence
CDS
ATGGCAAACATTCCTGATTGCCCCATGAGCCTAAAATCAATTCAGCATTATTTGAAGACTGCAGCAGAACACGACAACCGAGATGTGGTTGTAGCATATTGGTGTCGCCTTCACGCTTTACAAACCGGCCTGAAAATAACTACCAAGAAAACCCCTGAAGAAACCAAAGTTTTAATGGCTCTTATGGACTGGCTTGAAGAAATAAAAAAATCTCATGTAGACAATGAAGCAATCAGCAATGATGTAGCAGCCCAGGCCCATCTTGAAAACTATGCACTGAAATTGTTTCTGTATGCTGACAAACAGGACAGGGAACAAAATTATGGAAAAAATGTGGTCAAGGCATTTTATACTGCGGGTATGATCTATGATGTTCTTACCACATTTGGTGAACTGACAGATGAAGCTGCTCAGAATAGAAAGTATGCTAAGTGGAAGGCTGCGTATATTCATAACTGCCTTAAGAATGGAGAGACACCCGTACCAGGTCCAATGCAGTCAGAAGATGATCAAGGCAATGCAGGGATGTCTAGTGGCAATCAAGAAGCACCTTCGGATCCACCTCAACCTACTGATTTCACAAGTAACCCTCCAGAAACACCTCCCATAGCACCCACAAGCTTCAATAGCTCGTTACCAGATCCAAATGCAGTATTAAGAGCTGCATCTCAATTACCCCCTGTTCCGTACACTCCAGATCCAAATCCTGGTGGTTTTGTGCCATATGACCCAACCCAACAACCTCAACCACCTCAGCCAACCACCTTTCATAGTGACACTACCAATTTGCCAACTCTTACACCTGATCAAATAGGAAAAGCTCAGAAATACTGTAAATGGGCAAGTAGTGCTTTGAATTATGATGATGTTAAAACTGCTATTTATAATCTTAAGAGTGCTCTGGAGTTACTGCAAACTGGACGTGATCCAGCCTGA
Protein
MANIPDCPMSLKSIQHYLKTAAEHDNRDVVVAYWCRLHALQTGLKITTKKTPEETKVLMALMDWLEEIKKSHVDNEAISNDVAAQAHLENYALKLFLYADKQDREQNYGKNVVKAFYTAGMIYDVLTTFGELTDEAAQNRKYAKWKAAYIHNCLKNGETPVPGPMQSEDDQGNAGMSSGNQEAPSDPPQPTDFTSNPPETPPIAPTSFNSSLPDPNAVLRAASQLPPVPYTPDPNPGGFVPYDPTQQPQPPQPTTFHSDTTNLPTLTPDQIGKAQKYCKWASSALNYDDVKTAIYNLKSALELLQTGRDPA
Summary
Uniprot
Q1HQ07
A0A2W1BIE8
A0A3S2P5C3
A0A2H1VC45
A0A194R0E6
A0A194Q3M6
+ More
A0A0L7LSU7 A0A1E1WLS9 S4PLU9 A0A212F0K1 A0A1Q3FC11 A0A1Q3FF18 B4J1X5 B3NES8 A0A1W4VVB5 A0A1A9Z3D0 A0A1A9Y9N0 A0A1B0B998 A0A1L8DW94 A0A182KRI7 A0A182I3G1 Q7QJJ1 A0A1L8DWR8 A0A084WNH4 A0A2J7QQT9 A0A0J9RNC1 Q9W0B3 D3TPI3 A0A1A9VYL9 B4PDR1 A0A023ENS2 A0A1B0DMG4 A0A182TTD4 B3M7R8 A0A182JQY0 Q16G25 A0A182X3K9 A0A0M4EJY6 A0A1S4G1Z9 W8BZF9 B4L973 Q2LZP7 B4H3G2 B4N713 A0A182VLI5 A0A182PH97 A0A182M0Z2 A0A0L0C7F5 A0A182WJ36 A0A067QSN9 A0A182XZ13 A0A2M4AQC2 A0A182J8G9 T1GT49 W5JH46 A0A0K8VWN1 A0A182Q682 A0A2M4BVC7 A0A2M4BUG7 A0A3B0J5K6 B4LBU0 A0A1I8MFK1 A0A1A9X2W6 A0A1I8PEM4 A0A1L8EIN9 A0A182FG13 A0A1L8EIT0 T1DPK8 A0A0K8TLF0 K7ITS7 A0A182NH60 A0A182RQU5 U5EZ32 B0WW61 A0A2M3ZAV2 A0A034VYC0 A0A0M8ZQ65 A0A336L7D9 A0A336MRR2 A0A2M3ZIV0 A0A1B6DCH3 A0A151XGJ9 A0A195F0P7 A0A195BZ24 A0A158P3M7 F4W4T4 A0A151J9Y5 B4QMI0 A0A069DYI2 A0A0A9Y937 A0A0K8SSV8 A0A1J1J3S8 A0A1B6GFC6 A0A195CT27 E9IK91 A0A023GCA8 A0A026WS13 A0A2R5LAF7 A0A0P4VT76 A0A023F2E0
A0A0L7LSU7 A0A1E1WLS9 S4PLU9 A0A212F0K1 A0A1Q3FC11 A0A1Q3FF18 B4J1X5 B3NES8 A0A1W4VVB5 A0A1A9Z3D0 A0A1A9Y9N0 A0A1B0B998 A0A1L8DW94 A0A182KRI7 A0A182I3G1 Q7QJJ1 A0A1L8DWR8 A0A084WNH4 A0A2J7QQT9 A0A0J9RNC1 Q9W0B3 D3TPI3 A0A1A9VYL9 B4PDR1 A0A023ENS2 A0A1B0DMG4 A0A182TTD4 B3M7R8 A0A182JQY0 Q16G25 A0A182X3K9 A0A0M4EJY6 A0A1S4G1Z9 W8BZF9 B4L973 Q2LZP7 B4H3G2 B4N713 A0A182VLI5 A0A182PH97 A0A182M0Z2 A0A0L0C7F5 A0A182WJ36 A0A067QSN9 A0A182XZ13 A0A2M4AQC2 A0A182J8G9 T1GT49 W5JH46 A0A0K8VWN1 A0A182Q682 A0A2M4BVC7 A0A2M4BUG7 A0A3B0J5K6 B4LBU0 A0A1I8MFK1 A0A1A9X2W6 A0A1I8PEM4 A0A1L8EIN9 A0A182FG13 A0A1L8EIT0 T1DPK8 A0A0K8TLF0 K7ITS7 A0A182NH60 A0A182RQU5 U5EZ32 B0WW61 A0A2M3ZAV2 A0A034VYC0 A0A0M8ZQ65 A0A336L7D9 A0A336MRR2 A0A2M3ZIV0 A0A1B6DCH3 A0A151XGJ9 A0A195F0P7 A0A195BZ24 A0A158P3M7 F4W4T4 A0A151J9Y5 B4QMI0 A0A069DYI2 A0A0A9Y937 A0A0K8SSV8 A0A1J1J3S8 A0A1B6GFC6 A0A195CT27 E9IK91 A0A023GCA8 A0A026WS13 A0A2R5LAF7 A0A0P4VT76 A0A023F2E0
Pubmed
19121390
28756777
26354079
26227816
23622113
22118469
+ More
17994087 20966253 12364791 14747013 17210077 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 17550304 24945155 17510324 24495485 15632085 26108605 24845553 25244985 20920257 23761445 25315136 26369729 20075255 25348373 21347285 21719571 26334808 25401762 26823975 21282665 24508170 27129103 25474469
17994087 20966253 12364791 14747013 17210077 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 17550304 24945155 17510324 24495485 15632085 26108605 24845553 25244985 20920257 23761445 25315136 26369729 20075255 25348373 21347285 21719571 26334808 25401762 26823975 21282665 24508170 27129103 25474469
EMBL
BABH01037021
DQ443245
JX569220
ABF51334.1
AFW03815.1
KZ150103
+ More
PZC73464.1 RSAL01000018 RVE52802.1 ODYU01001764 SOQ38420.1 KQ460883 KPJ11263.1 KQ459586 KPI98015.1 JTDY01000159 KOB78527.1 GDQN01003243 JAT87811.1 GAIX01001702 JAA90858.1 AGBW02011073 OWR47260.1 GFDL01009931 JAV25114.1 GFDL01008902 JAV26143.1 CH916366 EDV95900.1 CH954178 EDV50201.1 JXJN01010337 GFDF01003378 JAV10706.1 APCN01000208 AAAB01008807 EAA04380.2 GFDF01003379 JAV10705.1 ATLV01024604 KE525352 KFB51768.1 NEVH01012082 PNF30950.1 CM002912 KMY96904.1 AE014296 AY051549 AY094954 AAF47540.1 AAK92973.1 AAM11307.1 CCAG010023908 EZ423335 ADD19611.1 CM000159 EDW92876.1 GAPW01002516 JAC11082.1 AJVK01001320 CH902618 EDV38791.1 CH478342 EAT33190.1 CP012525 ALC44401.1 GAMC01011932 GAMC01011931 JAB94624.1 CH933816 EDW17248.1 CH379069 EAL29460.1 CH479206 EDW30902.1 CH964168 EDW80152.1 AXCM01004497 JRES01000810 KNC28205.1 KK853069 KDR11827.1 GGFK01009648 MBW42969.1 CAQQ02390717 ADMH02001258 ETN63386.1 GDHF01009027 JAI43287.1 AXCN02000877 GGFJ01007567 MBW56708.1 GGFJ01007558 MBW56699.1 OUUW01000002 SPP76965.1 CH940647 EDW68717.1 GFDG01000232 JAV18567.1 GFDG01000231 JAV18568.1 GAMD01003264 JAA98326.1 GDAI01002617 JAI14986.1 GANO01001520 JAB58351.1 DS232139 EDS35893.1 GGFM01004870 MBW25621.1 GAKP01010661 JAC48291.1 KQ435989 KOX67716.1 UFQS01002298 UFQT01002298 SSX13718.1 SSX33139.1 UFQT01001051 SSX28638.1 GGFM01007678 MBW28429.1 GEDC01013914 JAS23384.1 KQ982174 KYQ59340.1 KQ981880 KYN33936.1 KQ976394 KYM93168.1 ADTU01008268 GL887553 EGI70803.1 KQ979347 KYN21875.1 CM000363 EDX08837.1 GBGD01002335 JAC86554.1 GBHO01014895 GDHC01013198 JAG28709.1 JAQ05431.1 GBRD01009553 JAG56271.1 CVRI01000067 CRL06428.1 GECZ01008632 JAS61137.1 KQ977304 KYN03800.1 GL763924 EFZ19025.1 GBBM01003926 JAC31492.1 KK107119 EZA58733.1 GGLE01002334 MBY06460.1 GDKW01000174 JAI56421.1 GBBI01003262 JAC15450.1
PZC73464.1 RSAL01000018 RVE52802.1 ODYU01001764 SOQ38420.1 KQ460883 KPJ11263.1 KQ459586 KPI98015.1 JTDY01000159 KOB78527.1 GDQN01003243 JAT87811.1 GAIX01001702 JAA90858.1 AGBW02011073 OWR47260.1 GFDL01009931 JAV25114.1 GFDL01008902 JAV26143.1 CH916366 EDV95900.1 CH954178 EDV50201.1 JXJN01010337 GFDF01003378 JAV10706.1 APCN01000208 AAAB01008807 EAA04380.2 GFDF01003379 JAV10705.1 ATLV01024604 KE525352 KFB51768.1 NEVH01012082 PNF30950.1 CM002912 KMY96904.1 AE014296 AY051549 AY094954 AAF47540.1 AAK92973.1 AAM11307.1 CCAG010023908 EZ423335 ADD19611.1 CM000159 EDW92876.1 GAPW01002516 JAC11082.1 AJVK01001320 CH902618 EDV38791.1 CH478342 EAT33190.1 CP012525 ALC44401.1 GAMC01011932 GAMC01011931 JAB94624.1 CH933816 EDW17248.1 CH379069 EAL29460.1 CH479206 EDW30902.1 CH964168 EDW80152.1 AXCM01004497 JRES01000810 KNC28205.1 KK853069 KDR11827.1 GGFK01009648 MBW42969.1 CAQQ02390717 ADMH02001258 ETN63386.1 GDHF01009027 JAI43287.1 AXCN02000877 GGFJ01007567 MBW56708.1 GGFJ01007558 MBW56699.1 OUUW01000002 SPP76965.1 CH940647 EDW68717.1 GFDG01000232 JAV18567.1 GFDG01000231 JAV18568.1 GAMD01003264 JAA98326.1 GDAI01002617 JAI14986.1 GANO01001520 JAB58351.1 DS232139 EDS35893.1 GGFM01004870 MBW25621.1 GAKP01010661 JAC48291.1 KQ435989 KOX67716.1 UFQS01002298 UFQT01002298 SSX13718.1 SSX33139.1 UFQT01001051 SSX28638.1 GGFM01007678 MBW28429.1 GEDC01013914 JAS23384.1 KQ982174 KYQ59340.1 KQ981880 KYN33936.1 KQ976394 KYM93168.1 ADTU01008268 GL887553 EGI70803.1 KQ979347 KYN21875.1 CM000363 EDX08837.1 GBGD01002335 JAC86554.1 GBHO01014895 GDHC01013198 JAG28709.1 JAQ05431.1 GBRD01009553 JAG56271.1 CVRI01000067 CRL06428.1 GECZ01008632 JAS61137.1 KQ977304 KYN03800.1 GL763924 EFZ19025.1 GBBM01003926 JAC31492.1 KK107119 EZA58733.1 GGLE01002334 MBY06460.1 GDKW01000174 JAI56421.1 GBBI01003262 JAC15450.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000001070 UP000008711 UP000192221 UP000092445 UP000092443 UP000092460 UP000075882 UP000075840 UP000007062 UP000030765 UP000235965 UP000000803 UP000092444 UP000078200 UP000002282 UP000092462 UP000075902 UP000007801 UP000075881 UP000008820 UP000076407 UP000092553 UP000009192 UP000001819 UP000008744 UP000007798 UP000075903 UP000075885 UP000075883 UP000037069 UP000075920 UP000027135 UP000076408 UP000075880 UP000015102 UP000000673 UP000075886 UP000268350 UP000008792 UP000095301 UP000091820 UP000095300 UP000069272 UP000002358 UP000075884 UP000075900 UP000002320 UP000053105 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000078492 UP000000304 UP000183832 UP000078542 UP000053097
UP000001070 UP000008711 UP000192221 UP000092445 UP000092443 UP000092460 UP000075882 UP000075840 UP000007062 UP000030765 UP000235965 UP000000803 UP000092444 UP000078200 UP000002282 UP000092462 UP000075902 UP000007801 UP000075881 UP000008820 UP000076407 UP000092553 UP000009192 UP000001819 UP000008744 UP000007798 UP000075903 UP000075885 UP000075883 UP000037069 UP000075920 UP000027135 UP000076408 UP000075880 UP000015102 UP000000673 UP000075886 UP000268350 UP000008792 UP000095301 UP000091820 UP000095300 UP000069272 UP000002358 UP000075884 UP000075900 UP000002320 UP000053105 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000078492 UP000000304 UP000183832 UP000078542 UP000053097
Gene 3D
ProteinModelPortal
Q1HQ07
A0A2W1BIE8
A0A3S2P5C3
A0A2H1VC45
A0A194R0E6
A0A194Q3M6
+ More
A0A0L7LSU7 A0A1E1WLS9 S4PLU9 A0A212F0K1 A0A1Q3FC11 A0A1Q3FF18 B4J1X5 B3NES8 A0A1W4VVB5 A0A1A9Z3D0 A0A1A9Y9N0 A0A1B0B998 A0A1L8DW94 A0A182KRI7 A0A182I3G1 Q7QJJ1 A0A1L8DWR8 A0A084WNH4 A0A2J7QQT9 A0A0J9RNC1 Q9W0B3 D3TPI3 A0A1A9VYL9 B4PDR1 A0A023ENS2 A0A1B0DMG4 A0A182TTD4 B3M7R8 A0A182JQY0 Q16G25 A0A182X3K9 A0A0M4EJY6 A0A1S4G1Z9 W8BZF9 B4L973 Q2LZP7 B4H3G2 B4N713 A0A182VLI5 A0A182PH97 A0A182M0Z2 A0A0L0C7F5 A0A182WJ36 A0A067QSN9 A0A182XZ13 A0A2M4AQC2 A0A182J8G9 T1GT49 W5JH46 A0A0K8VWN1 A0A182Q682 A0A2M4BVC7 A0A2M4BUG7 A0A3B0J5K6 B4LBU0 A0A1I8MFK1 A0A1A9X2W6 A0A1I8PEM4 A0A1L8EIN9 A0A182FG13 A0A1L8EIT0 T1DPK8 A0A0K8TLF0 K7ITS7 A0A182NH60 A0A182RQU5 U5EZ32 B0WW61 A0A2M3ZAV2 A0A034VYC0 A0A0M8ZQ65 A0A336L7D9 A0A336MRR2 A0A2M3ZIV0 A0A1B6DCH3 A0A151XGJ9 A0A195F0P7 A0A195BZ24 A0A158P3M7 F4W4T4 A0A151J9Y5 B4QMI0 A0A069DYI2 A0A0A9Y937 A0A0K8SSV8 A0A1J1J3S8 A0A1B6GFC6 A0A195CT27 E9IK91 A0A023GCA8 A0A026WS13 A0A2R5LAF7 A0A0P4VT76 A0A023F2E0
A0A0L7LSU7 A0A1E1WLS9 S4PLU9 A0A212F0K1 A0A1Q3FC11 A0A1Q3FF18 B4J1X5 B3NES8 A0A1W4VVB5 A0A1A9Z3D0 A0A1A9Y9N0 A0A1B0B998 A0A1L8DW94 A0A182KRI7 A0A182I3G1 Q7QJJ1 A0A1L8DWR8 A0A084WNH4 A0A2J7QQT9 A0A0J9RNC1 Q9W0B3 D3TPI3 A0A1A9VYL9 B4PDR1 A0A023ENS2 A0A1B0DMG4 A0A182TTD4 B3M7R8 A0A182JQY0 Q16G25 A0A182X3K9 A0A0M4EJY6 A0A1S4G1Z9 W8BZF9 B4L973 Q2LZP7 B4H3G2 B4N713 A0A182VLI5 A0A182PH97 A0A182M0Z2 A0A0L0C7F5 A0A182WJ36 A0A067QSN9 A0A182XZ13 A0A2M4AQC2 A0A182J8G9 T1GT49 W5JH46 A0A0K8VWN1 A0A182Q682 A0A2M4BVC7 A0A2M4BUG7 A0A3B0J5K6 B4LBU0 A0A1I8MFK1 A0A1A9X2W6 A0A1I8PEM4 A0A1L8EIN9 A0A182FG13 A0A1L8EIT0 T1DPK8 A0A0K8TLF0 K7ITS7 A0A182NH60 A0A182RQU5 U5EZ32 B0WW61 A0A2M3ZAV2 A0A034VYC0 A0A0M8ZQ65 A0A336L7D9 A0A336MRR2 A0A2M3ZIV0 A0A1B6DCH3 A0A151XGJ9 A0A195F0P7 A0A195BZ24 A0A158P3M7 F4W4T4 A0A151J9Y5 B4QMI0 A0A069DYI2 A0A0A9Y937 A0A0K8SSV8 A0A1J1J3S8 A0A1B6GFC6 A0A195CT27 E9IK91 A0A023GCA8 A0A026WS13 A0A2R5LAF7 A0A0P4VT76 A0A023F2E0
PDB
2LXM
E-value=1.37921e-49,
Score=494
Ontologies
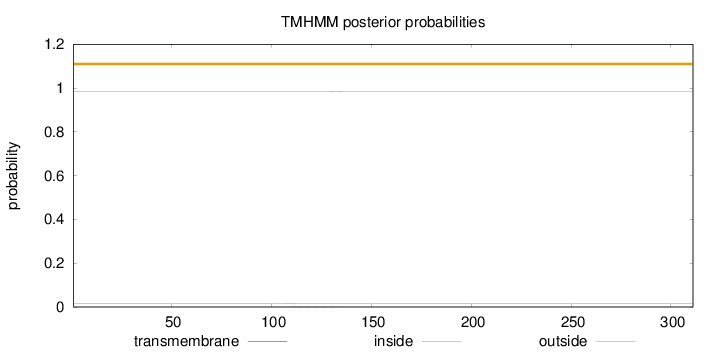
Topology
Length:
311
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04724
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.01686
outside
1 - 311
Population Genetic Test Statistics
Pi
240.384338
Theta
190.895638
Tajima's D
0.732529
CLR
0.122563
CSRT
0.576871156442178
Interpretation
Uncertain
