Gene
KWMTBOMO13293 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004691
Annotation
parp_[Bombyx_mori]
Full name
Poly [ADP-ribose] polymerase
Alternative Name
NAD(+) ADP-ribosyltransferase
Poly[ADP-ribose] synthase
Protein ADP-ribosyltransferase
Protein ADP-ribosyltransferase Parp
Poly[ADP-ribose] synthase
Protein ADP-ribosyltransferase
Protein ADP-ribosyltransferase Parp
Location in the cell
Nuclear Reliability : 2.709
Sequence
CDS
ATGGCAGACTTGCCATATCAAGTTGAATACGCAAAGACAGGTAGAGCAGCCTGTAAGGGTTGTAAACAAAAAATTGATCAAGGCTCACTGCGCATTGCTGTGATGGTACAGTCAGCTTTCTTTGATGGAAAACAACCAAACTGGCATCATGAAGATTGTTTTTTTAAAAAGCGACGTTTAAACAGTTTCACTGAAATTGCAAACTTCAATGTACTTAAAAATGAAGACCAGAAAAGAATAAAAACTACTATAGAAAACAGTGCTGGTGCTGTTGTAATGCCCATTCAAGAAACCAAAACCAAAGGCAAAGGTAAACGTAACAAGAGAGCCGCCCCAGAATCAGATAACTCTGCGCTGAAGGACTTCAAAGTTGAATATTCAAAAAGCAGTAGGGCCACGTGTCCTGAATGTGAAATTAAAATTTGTAAGGATGAAATTAGAATATGTAAGATTTTATATGATACAGAAGTTGGTATGAAATATGGTGGACAACCAAGATGGCACCACTTGCCTTGTTTCGTTAAATGCAGAAATGAACTTCTGTACTTCGCTGGAGGAGAGAATCTACCAGGATTTGATGATCTTAAGAAAGCAGATCAGATAATTGTGAAAACGGAGATAAAACCATTTGAAAGCCCTGACATTCCAATAAAAAAATTGAAGAACGAAGTAAAAGACGAAAAAGAGTTGAAAGAGGACAAAGCACTTGAAAACAAAATTGAAAAACAAAACAAATTGTTTGATAAACATAGAAATGCTTTAATGAATACAAATAAAAATGATGTAGAAAAAATTTTGGAGTACAATTTACAGGGAATCCCAACAGGAAAGACTGAGTGTTTTGATCGTTTAGCAGATTGTATGACATTCGGAGCTCTCGAGGCCTGTCCTGAATGTAAACAAGGGCAGTTAGTTTTAGATACCCTTGGCTATAAATGCACAGGTAATTTGAGCGAGTGGACCAAATGTACCTATCAAACTGATAAACCGAAGAGGAGGGCTCTGAAAATACCAGAAAAATACAAAGAGCTTGATTGTTTCCGCTCGTTTAAGCCCACGGTCCGCACTCGTTTGTTCCACGCTCCACTTCCATCACAAAACTTTGTTGTGAAACAAGAAGAGACAGAAGTAAAAAAAGAAATCCGTGTGCCGCCTTTGAAAAACTTGAAATTCTTCATTTACGGAACATTGAAGAAGAATGAAAAGGAGAATATCAAGAATCGCATTATCAAGCTCGGAGGAGACCCCGTCAGTAAACTGTCTGAAGCTACGGTCGCAGTCATCTCTACTAAAGCGGATGTCGAGAAAATGTCTTCTAAAATGCAATCCATACAATATCTAGATATTGTAGTTCTAGATGAAAGTTTTCTAAATTCGATTGACCCTGAGACCGGAACTGTAGCCAGATCCCTGGAGTTAATGAAAGAATATAATATCGCCGATTGGGGTGTTGATCCTGCCTCTAAGCTGCCCCAAGATATTATTGATGGAAAATCGATACCCAAGAGTAAAAGCATGTATGTCAAGTCTGAAAAATCAGCAGTCAAACTCAAGATCCGCGACGGCAACGCTGTGGACCCGGCATCGGAACTGGACGAGGAGTGCCACGTGTACACGGACCGCTACAAAACAACGTATTCGGTCGTGCTCTCGCTCACTGATGTTGTCGCTCAGAAGAACTCCTTCTACAAACTGCAGATCTTGAAACATAATAAAGAGAACAGATACTGGTTGTTCCGTTCGTGGGGTCGGATCGGCACAACGATCGGCGGCAGTAAAGTCGAGAAGATGAAAGATCTCGATAACGCTTTGGACAGATTCCAAGCGCTGTACGAAGAAAAAACTGAGAACCGTTGGAAAGACAGGAATAACTTTGTCAAGATTCCGGGGCGATACTTCCCGCTGGAAATAGACTACGGTGACAATGAGACGAAAACACTCACTGTCGACCCCAAGTGTGAGCTCGCCGAGCCCGTGCAGCGACTCATCGTCACCATCTTTGACATCCACTACATGAAGCAGACCTTGCTCGAATTCGAGTTGGACACAGAGAAAATGCCTCTCGGAAAGTTGTCTAAGAAACAAATAAAAGCAGGCTACAAAGTATTATCTGAACTGCAGAATCTCATAAAAAGCGGAGATGCGAGTCGAAATAAAATTGTGGATGCCACTAATAGATTTTTCACTTTAATACCCCACGACTTCGGGGTAAATAACGCACCGCTGTTGGATAATAATGAAATTATAAAGACGAAGACCGATATGATGGATAACCTGTTGGAAATTGAAATTGCGTATGGTCTATTACAATCAGACACTGACGACACGGTGAGCCCCATCGAAGGCCACTACAAGAAGCTGAAGGCGGAGATAGCGCCGCTCGACAAAACCACCGACGAGTTCGAAATGATCCTGGAGTACGTGCGGAACACGCACGGAAAAACGCACAACTTCTACACGCTCAATGTGGAAGAGGTGTTCAAAGTTATCCGCGAGGGAGAAGATAAGCGTTACAAGCCCTTCAAGAAACTCCACAACCGCCGCCTCTTGTGGCACGGCTCCCGCACCACCAACTTCGCCGGCATTATTTCACAAGGTCTTCGTATTGCGCCGCCCGAAGCGCCGGTCACGGGCTACATGTTCGGCAAGGGCATATACTTTGCGGACATGGTCTCCAAGTCCGCCAACTACTGCTGCACCACCAGAAACAACCCCGTCGGCCTCATGTTGCTGTGTGAGGTCGCTCTCGGGAATATGAAGGAGTGTGTCAATGCTGAAGGTTTCTCTAAAGCCCCGTCCGGCACGCATTCCGTGTGGGGCGTCGGTCGAACTGAGCCGGACCCGAAAATGAATAAGGAATTGCCCGACGGACTCATTGTGCCGCTCGGGACGCCCGTCGACAGGGCGGAGAACAGCACGCTGCTGTACAACGAGTTCATCGTGTACGACGTGGCTCAGGTCAAAGTGAAATACCTGATTGAAATGAGATTCGATTACAGATAA
Protein
MADLPYQVEYAKTGRAACKGCKQKIDQGSLRIAVMVQSAFFDGKQPNWHHEDCFFKKRRLNSFTEIANFNVLKNEDQKRIKTTIENSAGAVVMPIQETKTKGKGKRNKRAAPESDNSALKDFKVEYSKSSRATCPECEIKICKDEIRICKILYDTEVGMKYGGQPRWHHLPCFVKCRNELLYFAGGENLPGFDDLKKADQIIVKTEIKPFESPDIPIKKLKNEVKDEKELKEDKALENKIEKQNKLFDKHRNALMNTNKNDVEKILEYNLQGIPTGKTECFDRLADCMTFGALEACPECKQGQLVLDTLGYKCTGNLSEWTKCTYQTDKPKRRALKIPEKYKELDCFRSFKPTVRTRLFHAPLPSQNFVVKQEETEVKKEIRVPPLKNLKFFIYGTLKKNEKENIKNRIIKLGGDPVSKLSEATVAVISTKADVEKMSSKMQSIQYLDIVVLDESFLNSIDPETGTVARSLELMKEYNIADWGVDPASKLPQDIIDGKSIPKSKSMYVKSEKSAVKLKIRDGNAVDPASELDEECHVYTDRYKTTYSVVLSLTDVVAQKNSFYKLQILKHNKENRYWLFRSWGRIGTTIGGSKVEKMKDLDNALDRFQALYEEKTENRWKDRNNFVKIPGRYFPLEIDYGDNETKTLTVDPKCELAEPVQRLIVTIFDIHYMKQTLLEFELDTEKMPLGKLSKKQIKAGYKVLSELQNLIKSGDASRNKIVDATNRFFTLIPHDFGVNNAPLLDNNEIIKTKTDMMDNLLEIEIAYGLLQSDTDDTVSPIEGHYKKLKAEIAPLDKTTDEFEMILEYVRNTHGKTHNFYTLNVEEVFKVIREGEDKRYKPFKKLHNRRLLWHGSRTTNFAGIISQGLRIAPPEAPVTGYMFGKGIYFADMVSKSANYCCTTRNNPVGLMLLCEVALGNMKECVNAEGFSKAPSGTHSVWGVGRTEPDPKMNKELPDGLIVPLGTPVDRAENSTLLYNEFIVYDVAQVKVKYLIEMRFDYR
Summary
Description
Poly-ADP-ribosyltransferase that mediates poly-ADP-ribosylation of proteins and plays a key role in DNA repair. Mainly mediates glutamate and aspartate ADP-ribosylation of target proteins: the ADP-D-ribosyl group of NAD(+) is transferred to the acceptor carboxyl group of glutamate and aspartate residues and further ADP-ribosyl groups are transferred to the 2'-position of the terminal adenosine moiety, building up a polymer with an average chain length of 20-30 units.
Isoform e: Plays a fundamental role in organizing chromatin on a global scale. Autoregulates Parp transcription by influencing the chromatin structure of its heterochromatic environment.
Isoform e: Plays a fundamental role in organizing chromatin on a global scale. Autoregulates Parp transcription by influencing the chromatin structure of its heterochromatic environment.
Catalytic Activity
NAD(+) + (ADP-D-ribosyl)(n)-acceptor = nicotinamide + (ADP-D-ribosyl)(n+1)-acceptor.
L-aspartyl-[protein] + NAD(+) = 4-O-(ADP-D-ribosyl)-L-aspartyl-[protein] + nicotinamide
L-glutamyl-[protein] + NAD(+) = 5-O-(ADP-D-ribosyl)-L-glutamyl-[protein] + nicotinamide
L-aspartyl-[protein] + NAD(+) = 4-O-(ADP-D-ribosyl)-L-aspartyl-[protein] + nicotinamide
L-glutamyl-[protein] + NAD(+) = 5-O-(ADP-D-ribosyl)-L-glutamyl-[protein] + nicotinamide
Miscellaneous
The ADP-D-ribosyl group of NAD(+) is transferred to an acceptor carboxyl group on a histone or the enzyme itself, and further ADP-ribosyl groups are transferred to the 2'-position of the terminal adenosine moiety, building up a polymer with an average chain length of 20-30 units.
Keywords
ADP-ribosylation
Direct protein sequencing
DNA-binding
Glycosyltransferase
Metal-binding
NAD
Nucleus
Repeat
Transferase
Zinc
Zinc-finger
Alternative splicing
Chromatin regulator
Complete proteome
Reference proteome
Feature
chain Poly [ADP-ribose] polymerase
splice variant In isoform I.
splice variant In isoform I.
Uniprot
E9JEI6
A0A212F0J8
A0A2H1VJQ4
A0A194PYA6
A0A2H1VI59
A0A2J7PXX8
+ More
A0A2J7PXW9 A0A336MUR8 A0A2Z5U1X2 U5EZI1 A0A1L8DH70 A0A1A9XX79 A0A194PHW5 A0A1Y1KWX6 A0A1W4XRS2 A0A1B0BYD9 A0A1I8PWS2 A0A1A9WQP6 B4K4G8 A0A1B0GBV0 A0A1A9Z7M1 A0A154P952 A0A034V6Q0 Q11208 A0A1I8NDN9 A0A0A1X5Z5 A0A069DZP0 A0A0A9YXP0 Q16NZ4 B4JYU1 W5J9L6 T1IQQ9 A0A0M9A7Q7 A0A0T6B3Q7 A0A088A5D3 A0A182G2R9 A0A0K8U6G2 A0A182YM37 A0A0V0G612 A0A182FSK4 A0A3B0KIL8 B4GMA4 A0A2A3E5X0 E2AAA6 E2BH34 A0A232EXK9 A0A1S3JN87 A0A182JNU7 B0WYS5 A0A084WSM5 E9IXR5 A0A0Q5U9S9 A0A1Q3EXW2 K7IV59 B4MC98 W8BF08 F4X1H8 A0A0L7R840 A0A0P4W0D4 W8BJP9 A0A182R0F8 A0A195DLT0 A0A182VQR6 A0A195FRQ8 A0A194QPL8 A0A182N598 A0A151X7L1 P35875 A0A0C9REF5 A0A182RIX6 A0A182MGH8 A0A0J7L6S8 A0A158NA68 A0A195BKD2 A0A182PE54 A0A310S7N4 A0A0P9BT56 B4NZE0 A0A182HJB0 A0A026WPG3 A0A3L8D5Q5 A0A182V5X3 A0A182LCH6 T1P821 A0A182TIM5 K1QLU6 C8CCN1 T1HMS8 A0A0K2SYF4 A0A182XE12 Q7QBC7 A0A1B6DFG5 A0A0P8Y6K9 A0A0P5HT38 A0A2P8ZDR5 A0A0P6G395 A0A0P6EZW2 A0A0P5R332 A0A0P5IJS6 A0A164YKT3
A0A2J7PXW9 A0A336MUR8 A0A2Z5U1X2 U5EZI1 A0A1L8DH70 A0A1A9XX79 A0A194PHW5 A0A1Y1KWX6 A0A1W4XRS2 A0A1B0BYD9 A0A1I8PWS2 A0A1A9WQP6 B4K4G8 A0A1B0GBV0 A0A1A9Z7M1 A0A154P952 A0A034V6Q0 Q11208 A0A1I8NDN9 A0A0A1X5Z5 A0A069DZP0 A0A0A9YXP0 Q16NZ4 B4JYU1 W5J9L6 T1IQQ9 A0A0M9A7Q7 A0A0T6B3Q7 A0A088A5D3 A0A182G2R9 A0A0K8U6G2 A0A182YM37 A0A0V0G612 A0A182FSK4 A0A3B0KIL8 B4GMA4 A0A2A3E5X0 E2AAA6 E2BH34 A0A232EXK9 A0A1S3JN87 A0A182JNU7 B0WYS5 A0A084WSM5 E9IXR5 A0A0Q5U9S9 A0A1Q3EXW2 K7IV59 B4MC98 W8BF08 F4X1H8 A0A0L7R840 A0A0P4W0D4 W8BJP9 A0A182R0F8 A0A195DLT0 A0A182VQR6 A0A195FRQ8 A0A194QPL8 A0A182N598 A0A151X7L1 P35875 A0A0C9REF5 A0A182RIX6 A0A182MGH8 A0A0J7L6S8 A0A158NA68 A0A195BKD2 A0A182PE54 A0A310S7N4 A0A0P9BT56 B4NZE0 A0A182HJB0 A0A026WPG3 A0A3L8D5Q5 A0A182V5X3 A0A182LCH6 T1P821 A0A182TIM5 K1QLU6 C8CCN1 T1HMS8 A0A0K2SYF4 A0A182XE12 Q7QBC7 A0A1B6DFG5 A0A0P8Y6K9 A0A0P5HT38 A0A2P8ZDR5 A0A0P6G395 A0A0P6EZW2 A0A0P5R332 A0A0P5IJS6 A0A164YKT3
EC Number
2.4.2.-
2.4.2.30
2.4.2.30
Pubmed
19121390
21040523
22118469
26354079
26760975
28004739
+ More
17994087 25348373 8125121 25315136 25830018 26334808 25401762 26823975 17510324 20920257 23761445 26483478 25244985 20798317 28648823 24438588 21282665 20075255 24495485 21719571 8475096 9565614 12183365 10731132 12537572 12537569 21347285 17550304 24508170 30249741 20966253 22992520 19641118 12364791 29403074
17994087 25348373 8125121 25315136 25830018 26334808 25401762 26823975 17510324 20920257 23761445 26483478 25244985 20798317 28648823 24438588 21282665 20075255 24495485 21719571 8475096 9565614 12183365 10731132 12537572 12537569 21347285 17550304 24508170 30249741 20966253 22992520 19641118 12364791 29403074
EMBL
BABH01037034
BABH01037035
GQ426303
ADM32529.1
AGBW02011073
OWR47253.1
+ More
ODYU01002680 SOQ40484.1 KQ459586 KPI98008.1 SOQ40486.1 NEVH01020852 PNF21176.1 PNF21175.1 UFQT01001256 SSX29698.1 FX985744 BBA93631.1 GANO01000754 JAB59117.1 GFDF01008347 JAV05737.1 KQ459603 KPI92907.1 GEZM01071263 GEZM01071262 JAV65923.1 JXJN01022594 CH933806 EDW13920.1 CCAG010005890 KQ434846 KZC08363.1 GAKP01019946 JAC39006.1 D16482 GBXI01008132 JAD06160.1 GBGD01000305 JAC88584.1 GBHO01005832 GDHC01000572 JAG37772.1 JAQ18057.1 CH477801 EAT36080.1 CH916378 EDV98556.1 ADMH02002074 ETN59550.1 JH431312 KQ435729 KOX77633.1 LJIG01015963 KRT82026.1 JXUM01140010 KQ568989 KXJ68804.1 GDHF01030092 GDHF01003396 JAI22222.1 JAI48918.1 GECL01002996 JAP03128.1 OUUW01000014 SPP88360.1 CH479185 EDW37978.1 KZ288364 PBC26884.1 GL438057 EFN69632.1 GL448268 EFN84955.1 NNAY01001734 OXU23091.1 DS232197 EDS37185.1 ATLV01026624 KE525415 KFB53219.1 GL766762 EFZ14625.1 CH954178 KQS44468.1 GFDL01014896 JAV20149.1 AAZX01008502 CH940657 EDW63180.2 GAMC01009298 JAB97257.1 GL888529 EGI59706.1 KQ414633 KOC67050.1 GDRN01082474 GDRN01082473 JAI61826.1 GAMC01009297 JAB97258.1 AXCN02000148 KQ980734 KYN13833.1 KQ981305 KYN42972.1 KQ461194 KPJ06925.1 KQ982446 KYQ56319.1 D13806 AF051548 AF051544 AF051545 AF051546 AF051547 AF533701 AE014297 BT015238 AY118947 GBYB01005341 JAG75108.1 AXCM01001496 LBMM01000492 KMQ98224.1 ADTU01010050 KQ976455 KYM85147.1 KQ777311 OAD52149.1 CH902627 KPU74869.1 CM000157 EDW89853.1 APCN01002135 KK107153 EZA56984.1 QOIP01000012 RLU15812.1 KA644786 AFP59415.1 JH816283 EKC34863.1 GQ389638 ACU83597.1 ACPB03017427 HACA01001209 CDW18570.1 AAAB01008879 EAA08394.4 GEDC01012869 JAS24429.1 KPU74867.1 GDIQ01246178 JAK05547.1 PYGN01000086 PSN54639.1 GDIQ01041403 JAN53334.1 GDIQ01056248 JAN38489.1 GDIQ01108431 JAL43295.1 GDIQ01211538 JAK40187.1 LRGB01000872 KZS15355.1
ODYU01002680 SOQ40484.1 KQ459586 KPI98008.1 SOQ40486.1 NEVH01020852 PNF21176.1 PNF21175.1 UFQT01001256 SSX29698.1 FX985744 BBA93631.1 GANO01000754 JAB59117.1 GFDF01008347 JAV05737.1 KQ459603 KPI92907.1 GEZM01071263 GEZM01071262 JAV65923.1 JXJN01022594 CH933806 EDW13920.1 CCAG010005890 KQ434846 KZC08363.1 GAKP01019946 JAC39006.1 D16482 GBXI01008132 JAD06160.1 GBGD01000305 JAC88584.1 GBHO01005832 GDHC01000572 JAG37772.1 JAQ18057.1 CH477801 EAT36080.1 CH916378 EDV98556.1 ADMH02002074 ETN59550.1 JH431312 KQ435729 KOX77633.1 LJIG01015963 KRT82026.1 JXUM01140010 KQ568989 KXJ68804.1 GDHF01030092 GDHF01003396 JAI22222.1 JAI48918.1 GECL01002996 JAP03128.1 OUUW01000014 SPP88360.1 CH479185 EDW37978.1 KZ288364 PBC26884.1 GL438057 EFN69632.1 GL448268 EFN84955.1 NNAY01001734 OXU23091.1 DS232197 EDS37185.1 ATLV01026624 KE525415 KFB53219.1 GL766762 EFZ14625.1 CH954178 KQS44468.1 GFDL01014896 JAV20149.1 AAZX01008502 CH940657 EDW63180.2 GAMC01009298 JAB97257.1 GL888529 EGI59706.1 KQ414633 KOC67050.1 GDRN01082474 GDRN01082473 JAI61826.1 GAMC01009297 JAB97258.1 AXCN02000148 KQ980734 KYN13833.1 KQ981305 KYN42972.1 KQ461194 KPJ06925.1 KQ982446 KYQ56319.1 D13806 AF051548 AF051544 AF051545 AF051546 AF051547 AF533701 AE014297 BT015238 AY118947 GBYB01005341 JAG75108.1 AXCM01001496 LBMM01000492 KMQ98224.1 ADTU01010050 KQ976455 KYM85147.1 KQ777311 OAD52149.1 CH902627 KPU74869.1 CM000157 EDW89853.1 APCN01002135 KK107153 EZA56984.1 QOIP01000012 RLU15812.1 KA644786 AFP59415.1 JH816283 EKC34863.1 GQ389638 ACU83597.1 ACPB03017427 HACA01001209 CDW18570.1 AAAB01008879 EAA08394.4 GEDC01012869 JAS24429.1 KPU74867.1 GDIQ01246178 JAK05547.1 PYGN01000086 PSN54639.1 GDIQ01041403 JAN53334.1 GDIQ01056248 JAN38489.1 GDIQ01108431 JAL43295.1 GDIQ01211538 JAK40187.1 LRGB01000872 KZS15355.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000235965
UP000092443
UP000192223
+ More
UP000092460 UP000095300 UP000091820 UP000009192 UP000092444 UP000092445 UP000076502 UP000095301 UP000008820 UP000001070 UP000000673 UP000053105 UP000005203 UP000069940 UP000249989 UP000076408 UP000069272 UP000268350 UP000008744 UP000242457 UP000000311 UP000008237 UP000215335 UP000085678 UP000075881 UP000002320 UP000030765 UP000008711 UP000002358 UP000008792 UP000007755 UP000053825 UP000075886 UP000078492 UP000075920 UP000078541 UP000053240 UP000075884 UP000075809 UP000000803 UP000075900 UP000075883 UP000036403 UP000005205 UP000078540 UP000075885 UP000007801 UP000002282 UP000075840 UP000053097 UP000279307 UP000075903 UP000075882 UP000075902 UP000005408 UP000015103 UP000076407 UP000007062 UP000245037 UP000076858
UP000092460 UP000095300 UP000091820 UP000009192 UP000092444 UP000092445 UP000076502 UP000095301 UP000008820 UP000001070 UP000000673 UP000053105 UP000005203 UP000069940 UP000249989 UP000076408 UP000069272 UP000268350 UP000008744 UP000242457 UP000000311 UP000008237 UP000215335 UP000085678 UP000075881 UP000002320 UP000030765 UP000008711 UP000002358 UP000008792 UP000007755 UP000053825 UP000075886 UP000078492 UP000075920 UP000078541 UP000053240 UP000075884 UP000075809 UP000000803 UP000075900 UP000075883 UP000036403 UP000005205 UP000078540 UP000075885 UP000007801 UP000002282 UP000075840 UP000053097 UP000279307 UP000075903 UP000075882 UP000075902 UP000005408 UP000015103 UP000076407 UP000007062 UP000245037 UP000076858
Pfam
Interpro
IPR008893
WGR_domain
+ More
IPR001510 Znf_PARP
IPR004102 Poly(ADP-ribose)pol_reg_dom
IPR036420 BRCT_dom_sf
IPR001357 BRCT_dom
IPR012982 PADR1
IPR036616 Poly(ADP-ribose)pol_reg_dom_sf
IPR036957 Znf_PARP_sf
IPR038650 PADR1_dom_sf
IPR012317 Poly(ADP-ribose)pol_cat_dom
IPR036930 WGR_dom_sf
IPR008288 PARP
IPR001510 Znf_PARP
IPR004102 Poly(ADP-ribose)pol_reg_dom
IPR036420 BRCT_dom_sf
IPR001357 BRCT_dom
IPR012982 PADR1
IPR036616 Poly(ADP-ribose)pol_reg_dom_sf
IPR036957 Znf_PARP_sf
IPR038650 PADR1_dom_sf
IPR012317 Poly(ADP-ribose)pol_cat_dom
IPR036930 WGR_dom_sf
IPR008288 PARP
CDD
ProteinModelPortal
E9JEI6
A0A212F0J8
A0A2H1VJQ4
A0A194PYA6
A0A2H1VI59
A0A2J7PXX8
+ More
A0A2J7PXW9 A0A336MUR8 A0A2Z5U1X2 U5EZI1 A0A1L8DH70 A0A1A9XX79 A0A194PHW5 A0A1Y1KWX6 A0A1W4XRS2 A0A1B0BYD9 A0A1I8PWS2 A0A1A9WQP6 B4K4G8 A0A1B0GBV0 A0A1A9Z7M1 A0A154P952 A0A034V6Q0 Q11208 A0A1I8NDN9 A0A0A1X5Z5 A0A069DZP0 A0A0A9YXP0 Q16NZ4 B4JYU1 W5J9L6 T1IQQ9 A0A0M9A7Q7 A0A0T6B3Q7 A0A088A5D3 A0A182G2R9 A0A0K8U6G2 A0A182YM37 A0A0V0G612 A0A182FSK4 A0A3B0KIL8 B4GMA4 A0A2A3E5X0 E2AAA6 E2BH34 A0A232EXK9 A0A1S3JN87 A0A182JNU7 B0WYS5 A0A084WSM5 E9IXR5 A0A0Q5U9S9 A0A1Q3EXW2 K7IV59 B4MC98 W8BF08 F4X1H8 A0A0L7R840 A0A0P4W0D4 W8BJP9 A0A182R0F8 A0A195DLT0 A0A182VQR6 A0A195FRQ8 A0A194QPL8 A0A182N598 A0A151X7L1 P35875 A0A0C9REF5 A0A182RIX6 A0A182MGH8 A0A0J7L6S8 A0A158NA68 A0A195BKD2 A0A182PE54 A0A310S7N4 A0A0P9BT56 B4NZE0 A0A182HJB0 A0A026WPG3 A0A3L8D5Q5 A0A182V5X3 A0A182LCH6 T1P821 A0A182TIM5 K1QLU6 C8CCN1 T1HMS8 A0A0K2SYF4 A0A182XE12 Q7QBC7 A0A1B6DFG5 A0A0P8Y6K9 A0A0P5HT38 A0A2P8ZDR5 A0A0P6G395 A0A0P6EZW2 A0A0P5R332 A0A0P5IJS6 A0A164YKT3
A0A2J7PXW9 A0A336MUR8 A0A2Z5U1X2 U5EZI1 A0A1L8DH70 A0A1A9XX79 A0A194PHW5 A0A1Y1KWX6 A0A1W4XRS2 A0A1B0BYD9 A0A1I8PWS2 A0A1A9WQP6 B4K4G8 A0A1B0GBV0 A0A1A9Z7M1 A0A154P952 A0A034V6Q0 Q11208 A0A1I8NDN9 A0A0A1X5Z5 A0A069DZP0 A0A0A9YXP0 Q16NZ4 B4JYU1 W5J9L6 T1IQQ9 A0A0M9A7Q7 A0A0T6B3Q7 A0A088A5D3 A0A182G2R9 A0A0K8U6G2 A0A182YM37 A0A0V0G612 A0A182FSK4 A0A3B0KIL8 B4GMA4 A0A2A3E5X0 E2AAA6 E2BH34 A0A232EXK9 A0A1S3JN87 A0A182JNU7 B0WYS5 A0A084WSM5 E9IXR5 A0A0Q5U9S9 A0A1Q3EXW2 K7IV59 B4MC98 W8BF08 F4X1H8 A0A0L7R840 A0A0P4W0D4 W8BJP9 A0A182R0F8 A0A195DLT0 A0A182VQR6 A0A195FRQ8 A0A194QPL8 A0A182N598 A0A151X7L1 P35875 A0A0C9REF5 A0A182RIX6 A0A182MGH8 A0A0J7L6S8 A0A158NA68 A0A195BKD2 A0A182PE54 A0A310S7N4 A0A0P9BT56 B4NZE0 A0A182HJB0 A0A026WPG3 A0A3L8D5Q5 A0A182V5X3 A0A182LCH6 T1P821 A0A182TIM5 K1QLU6 C8CCN1 T1HMS8 A0A0K2SYF4 A0A182XE12 Q7QBC7 A0A1B6DFG5 A0A0P8Y6K9 A0A0P5HT38 A0A2P8ZDR5 A0A0P6G395 A0A0P6EZW2 A0A0P5R332 A0A0P5IJS6 A0A164YKT3
PDB
4DQY
E-value=2.79124e-161,
Score=1463
Ontologies
KEGG
PATHWAY
GO
GO:0008270
GO:0006471
GO:0051287
GO:0005634
GO:0003677
GO:0003950
GO:0043484
GO:0051276
GO:1900182
GO:0015030
GO:0035080
GO:0042393
GO:0005694
GO:0006325
GO:0051457
GO:0045087
GO:0005730
GO:0009303
GO:0006355
GO:0008069
GO:0035363
GO:0030576
GO:0005703
GO:0035079
GO:0007000
GO:0006963
GO:0005719
GO:0005700
GO:0007552
GO:0070212
GO:0006302
GO:1990404
GO:0048869
GO:0003676
GO:0016491
GO:0015930
Topology
Subcellular location
Nucleus
Nucleolus
Nucleolus
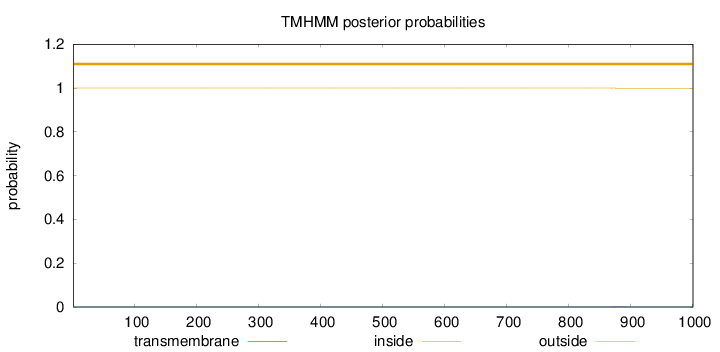
Length:
1000
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00985
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00002
outside
1 - 1000
Population Genetic Test Statistics
Pi
293.153927
Theta
198.496977
Tajima's D
3.419136
CLR
0.594162
CSRT
0.992200389980501
Interpretation
Uncertain
