Gene
KWMTBOMO13287 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004711
Annotation
PREDICTED:_ruvB-like_helicase_1_[Papilio_machaon]
Full name
RuvB-like helicase
+ More
RuvB-like helicase 1
DNA helicase
RuvB-like helicase 1
DNA helicase
Alternative Name
Pontin
Location in the cell
Cytoplasmic Reliability : 2.814
Sequence
CDS
ATGAAAATTGAAGAAGTGAAAAGCACAGCTAAAACGCAAAGGATCTCTGCTCACAGTCATATAAAAGGCTTGGGGTTGGATGAAAATGGTGTTCCTATTCAAATGGCAGCCGGTCTCGTGGGTCAAGAGTCTGCACGTGAGGCTGCAGGGATAGTTGTAGATATGATAAGAAGTAAGAAAATGGCCGGGCGAGCTTTACTCTTGGCAGGGCCTCCTGGTACTGGCAAAACTGCTATAGCTCTTGCCATCGCTCAGGAACTTGGAACTAAGGTTCCTTTCTGTCCAATGGTTGGTAGTGAAGTTTACAGCACAGAGATCAAGAAAACAGAGGTATTAATGGAGAATTTTCGACGTGCTATAGGCCTGCGTATACGTGAAACCAAAGAAGTATATGAGGGTGAAGTAACTGAGTTAACACCTGTTGAAACAGAAAATCCTGCTGGAGGCTATGGAAAAACAGTATCTCATGTGATCATAGGACTAAAAACGGCCAAGGGTACCAAACAACTTAAACTTGATCCAACTATATATGAATCGTTACAAAAGGAAAAGGTTGAAGTTGGGGATGTCATTTACATTGAAGCTAATTCGGGGGCAGTGAAGAGACAGGGTAGGAGTGATACTTTTGCTACTGAATTTGACCTTGAGGCTGAAGAATATGTTCCGCTACCCAAAGGTGATGTACACAAAAAGAAAGAAGTTGTACAAGATGTAACCTTACACGATTTGGATTGTGCTAATGCAAGACCGCAGGGAGGTCATGACATTATGTCTATGATGGGACAACTCATGAAACCAAAGAAGACTGAAATAACAGATAAACTTCGAAAAGAGATCAATAAAGTTGTGAACAAATACATAGATCAAGGGATTGCAGAACTGGTTCCTGGTGTGCTATTCATTGATGAGGTACACATGCTGGACATTGAAACTTTCACATACCTACATCGAGCGTTGGAGTCCGCGATAGCACCAATTGTTATATTTGCTACAAACAGAGGGCGTTGTCAAATCAGAGGGACAGAAGACATTGTATCACCACACGGCATTCCACTAGATCTATTAGATAGGTTATTAATAATACGCACTCTCCCGTATAACAAAGCAGAACTTTTACAGATATTAAAGCTCCGTGCGGTCACAGAAGGAATTTCAATCGAGGACGAAGCTTTGTCGGCGCTCTCTGAGATTGGAGCTCACACGACGCTCAGGTACGCAGTTCAGCTGCTAACTCCGGCGATGTTGGCCGCGCGAGCCGACGGCTGCCAGCGCATATCGCCGGCGCACGTGTCCTCGGTCCACACGCTGTTCCTTGACGCTAAAGCTTCAGCCAAAATCCTCACTCAACATTCCGATAAATACATGCAGTAA
Protein
MKIEEVKSTAKTQRISAHSHIKGLGLDENGVPIQMAAGLVGQESAREAAGIVVDMIRSKKMAGRALLLAGPPGTGKTAIALAIAQELGTKVPFCPMVGSEVYSTEIKKTEVLMENFRRAIGLRIRETKEVYEGEVTELTPVETENPAGGYGKTVSHVIIGLKTAKGTKQLKLDPTIYESLQKEKVEVGDVIYIEANSGAVKRQGRSDTFATEFDLEAEEYVPLPKGDVHKKKEVVQDVTLHDLDCANARPQGGHDIMSMMGQLMKPKKTEITDKLRKEINKVVNKYIDQGIAELVPGVLFIDEVHMLDIETFTYLHRALESAIAPIVIFATNRGRCQIRGTEDIVSPHGIPLDLLDRLLIIRTLPYNKAELLQILKLRAVTEGISIEDEALSALSEIGAHTTLRYAVQLLTPAMLAARADGCQRISPAHVSSVHTLFLDAKASAKILTQHSDKYMQ
Summary
Description
Proposed core component of the chromatin remodeling Ino80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Acts as a transcriptional coactivator in Wg signaling.
Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Acts as a transcriptional coactivator in Wg signaling.
Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Forms homohexameric rings. May form a dodecamer with rept made of two stacked hexameric rings. Component of the chromatin remodeling Ino80 complex (By similarity).
Similarity
Belongs to the RuvB family.
Keywords
Activator
ATP-binding
Cell cycle
Cell division
Chromatin regulator
Complete proteome
DNA damage
DNA recombination
DNA repair
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain RuvB-like helicase 1
Uniprot
H9J5C1
A0A2A4IVP2
A0A194R1Z9
A0A194Q3L6
A0A3S2NZ56
S4NSQ2
+ More
A0A212FAF2 A0A2H1WCW8 A0A067QQE5 A0A154PHZ2 A0A2J7QCA4 E9IDT2 E2B6Y7 A0A0J7NJC2 A0A195C2V5 A0A195BB51 A0A158NN53 A0A151IWH2 F4W8C7 A0A2A3E1P5 A0A087ZSJ5 A0A310SK01 A0A151X288 A0A195FAZ1 A0A1W4X1Z7 A0A1B6GJV2 A0A026WFT3 A0A1B6LL71 A0A1Y1LPU7 A0A232ESW5 A0A1B6HMU1 E0VMB1 K7J661 D6X0K0 A0A0C9QC64 A0A0N7Z9B2 R4FNT4 A0A0M8ZT85 A0A0V0G8C8 A0A069DU69 A0A210PMW5 A0A293MX40 A0A2R5LHA1 A0A1Z5LAI6 A0A182HCN0 U5EXK8 Q0IFL2 A0A2T7NVP9 T1IJL2 U4UF38 A0A1Q3F4B3 A0A1B0D5N8 A0A182XKF9 A0A182UB01 A0A2C9GQH2 A0A182VAW2 B0W8I1 W8CA62 W5JVX2 A0A0B7B8P4 L7MGZ0 A0A023FXK4 A0A084VTP9 A0A182XXI8 A0A131XCH3 A0A224Z0K8 A0A131Z1S1 A0A182PBG2 V4AYE4 A0A2R7X2A1 N6U3Y5 A0A023GNL1 G3MH90 A0A182IUZ6 A0A3F2Z113 A0A1B0CJ20 B7PYR8 A0A182MWY7 A0A2C9JD95 A0A1Y9HER1 A0A2M4ANB7 E9H9P0 A0A034WBU7 A0A224XQ29 A0A023FK87 T1FSY2 A0A131XXE0 A0A0K8W5T8 A0A182K3C8 A0A0P5XRN9 A0A267FV70 A0A1I8N241 A0A1J1HUP3 A0A182LCW5 A0A182FAX4 K4G9W9 A0A0T6B5H1 A0A0L0CDX9 A0A267F687 R7V1X6
A0A212FAF2 A0A2H1WCW8 A0A067QQE5 A0A154PHZ2 A0A2J7QCA4 E9IDT2 E2B6Y7 A0A0J7NJC2 A0A195C2V5 A0A195BB51 A0A158NN53 A0A151IWH2 F4W8C7 A0A2A3E1P5 A0A087ZSJ5 A0A310SK01 A0A151X288 A0A195FAZ1 A0A1W4X1Z7 A0A1B6GJV2 A0A026WFT3 A0A1B6LL71 A0A1Y1LPU7 A0A232ESW5 A0A1B6HMU1 E0VMB1 K7J661 D6X0K0 A0A0C9QC64 A0A0N7Z9B2 R4FNT4 A0A0M8ZT85 A0A0V0G8C8 A0A069DU69 A0A210PMW5 A0A293MX40 A0A2R5LHA1 A0A1Z5LAI6 A0A182HCN0 U5EXK8 Q0IFL2 A0A2T7NVP9 T1IJL2 U4UF38 A0A1Q3F4B3 A0A1B0D5N8 A0A182XKF9 A0A182UB01 A0A2C9GQH2 A0A182VAW2 B0W8I1 W8CA62 W5JVX2 A0A0B7B8P4 L7MGZ0 A0A023FXK4 A0A084VTP9 A0A182XXI8 A0A131XCH3 A0A224Z0K8 A0A131Z1S1 A0A182PBG2 V4AYE4 A0A2R7X2A1 N6U3Y5 A0A023GNL1 G3MH90 A0A182IUZ6 A0A3F2Z113 A0A1B0CJ20 B7PYR8 A0A182MWY7 A0A2C9JD95 A0A1Y9HER1 A0A2M4ANB7 E9H9P0 A0A034WBU7 A0A224XQ29 A0A023FK87 T1FSY2 A0A131XXE0 A0A0K8W5T8 A0A182K3C8 A0A0P5XRN9 A0A267FV70 A0A1I8N241 A0A1J1HUP3 A0A182LCW5 A0A182FAX4 K4G9W9 A0A0T6B5H1 A0A0L0CDX9 A0A267F687 R7V1X6
EC Number
3.6.4.12
Pubmed
19121390
26354079
23622113
22118469
24845553
21282665
+ More
20798317 21347285 21719571 24508170 30249741 28004739 28648823 20566863 20075255 18362917 19820115 27129103 26334808 28812685 28528879 26483478 17510324 23537049 24495485 20920257 23761445 25576852 24438588 25244985 28049606 28797301 26830274 23254933 22216098 15562597 21292972 25348373 25315136 20966253 23056606 26108605
20798317 21347285 21719571 24508170 30249741 28004739 28648823 20566863 20075255 18362917 19820115 27129103 26334808 28812685 28528879 26483478 17510324 23537049 24495485 20920257 23761445 25576852 24438588 25244985 28049606 28797301 26830274 23254933 22216098 15562597 21292972 25348373 25315136 20966253 23056606 26108605
EMBL
BABH01037043
NWSH01005962
PCG63791.1
KQ460883
KPJ11275.1
KQ459586
+ More
KPI98005.1 RSAL01000018 RVE52812.1 GAIX01013977 JAA78583.1 AGBW02009486 OWR50708.1 ODYU01007822 SOQ50913.1 KK853050 KDR12040.1 KQ434912 KZC11417.1 NEVH01016290 PNF26215.1 GL762535 EFZ21241.1 GL446071 EFN88529.1 LBMM01004290 KMQ92600.1 KQ978344 KYM94925.1 KQ976537 KYM81419.1 ADTU01021054 KQ980854 KYN12179.1 GL887908 EGI69418.1 KZ288446 PBC25620.1 KQ765330 OAD53965.1 KQ982579 KYQ54545.1 KQ981693 KYN37598.1 GECZ01016759 GECZ01007037 JAS53010.1 JAS62732.1 KK107263 QOIP01000011 EZA53889.1 RLU16931.1 GEBQ01015554 JAT24423.1 GEZM01050187 JAV75682.1 NNAY01002380 OXU21407.1 GECU01031740 JAS75966.1 DS235306 EEB14517.1 KQ971371 EFA10722.1 GBYB01000864 JAG70631.1 GDKW01001035 JAI55560.1 ACPB03010982 GAHY01000448 JAA77062.1 KQ435903 KOX69059.1 GECL01001713 JAP04411.1 GBGD01001419 JAC87470.1 NEDP02005581 OWF37766.1 GFWV01019182 MAA43910.1 GGLE01004669 MBY08795.1 GFJQ02002527 JAW04443.1 JXUM01127766 KQ567233 KXJ69542.1 GANO01000904 JAB58967.1 CH477312 EAT43892.1 PZQS01000009 PVD25245.1 JH430315 KB632294 ERL91652.1 GFDL01012686 JAV22359.1 AJVK01011851 APCN01002169 DS231859 EDS39031.1 GAMC01001554 JAC05002.1 ADMH02000300 ETN67049.1 HACG01041826 CEK88691.1 GACK01002501 JAA62533.1 GBBL01001101 JAC26219.1 ATLV01016383 KE525080 KFB41343.1 GEFH01004756 JAP63825.1 GFPF01011559 MAA22705.1 GEDV01004151 JAP84406.1 KB201224 ESO98661.1 KK856508 PTY25924.1 APGK01050244 KB741169 ENN73292.1 GBBM01000943 JAC34475.1 JO841241 AEO32858.1 AJWK01013975 ABJB010179771 ABJB010262550 ABJB010671266 DS821669 EEC11740.1 AXCM01004705 GGFK01008954 MBW42275.1 GL732609 EFX71513.1 GAKP01007709 JAC51243.1 GFTR01006315 JAW10111.1 GBBK01002450 JAC22032.1 AMQM01003889 KB096365 ESO05157.1 GEFM01003872 JAP71924.1 GDHF01005895 JAI46419.1 GDIP01068291 JAM35424.1 NIVC01000731 PAA77636.1 CVRI01000021 CRK91792.1 JX053104 JX210316 AFK11332.1 AFM88630.1 LJIG01009677 KRT82601.1 JRES01000501 KNC30618.1 NIVC01001336 PAA69268.1 AMQN01005385 KB295903 ELU12497.1
KPI98005.1 RSAL01000018 RVE52812.1 GAIX01013977 JAA78583.1 AGBW02009486 OWR50708.1 ODYU01007822 SOQ50913.1 KK853050 KDR12040.1 KQ434912 KZC11417.1 NEVH01016290 PNF26215.1 GL762535 EFZ21241.1 GL446071 EFN88529.1 LBMM01004290 KMQ92600.1 KQ978344 KYM94925.1 KQ976537 KYM81419.1 ADTU01021054 KQ980854 KYN12179.1 GL887908 EGI69418.1 KZ288446 PBC25620.1 KQ765330 OAD53965.1 KQ982579 KYQ54545.1 KQ981693 KYN37598.1 GECZ01016759 GECZ01007037 JAS53010.1 JAS62732.1 KK107263 QOIP01000011 EZA53889.1 RLU16931.1 GEBQ01015554 JAT24423.1 GEZM01050187 JAV75682.1 NNAY01002380 OXU21407.1 GECU01031740 JAS75966.1 DS235306 EEB14517.1 KQ971371 EFA10722.1 GBYB01000864 JAG70631.1 GDKW01001035 JAI55560.1 ACPB03010982 GAHY01000448 JAA77062.1 KQ435903 KOX69059.1 GECL01001713 JAP04411.1 GBGD01001419 JAC87470.1 NEDP02005581 OWF37766.1 GFWV01019182 MAA43910.1 GGLE01004669 MBY08795.1 GFJQ02002527 JAW04443.1 JXUM01127766 KQ567233 KXJ69542.1 GANO01000904 JAB58967.1 CH477312 EAT43892.1 PZQS01000009 PVD25245.1 JH430315 KB632294 ERL91652.1 GFDL01012686 JAV22359.1 AJVK01011851 APCN01002169 DS231859 EDS39031.1 GAMC01001554 JAC05002.1 ADMH02000300 ETN67049.1 HACG01041826 CEK88691.1 GACK01002501 JAA62533.1 GBBL01001101 JAC26219.1 ATLV01016383 KE525080 KFB41343.1 GEFH01004756 JAP63825.1 GFPF01011559 MAA22705.1 GEDV01004151 JAP84406.1 KB201224 ESO98661.1 KK856508 PTY25924.1 APGK01050244 KB741169 ENN73292.1 GBBM01000943 JAC34475.1 JO841241 AEO32858.1 AJWK01013975 ABJB010179771 ABJB010262550 ABJB010671266 DS821669 EEC11740.1 AXCM01004705 GGFK01008954 MBW42275.1 GL732609 EFX71513.1 GAKP01007709 JAC51243.1 GFTR01006315 JAW10111.1 GBBK01002450 JAC22032.1 AMQM01003889 KB096365 ESO05157.1 GEFM01003872 JAP71924.1 GDHF01005895 JAI46419.1 GDIP01068291 JAM35424.1 NIVC01000731 PAA77636.1 CVRI01000021 CRK91792.1 JX053104 JX210316 AFK11332.1 AFM88630.1 LJIG01009677 KRT82601.1 JRES01000501 KNC30618.1 NIVC01001336 PAA69268.1 AMQN01005385 KB295903 ELU12497.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000027135 UP000076502 UP000235965 UP000008237 UP000036403 UP000078542 UP000078540 UP000005205 UP000078492 UP000007755 UP000242457 UP000005203 UP000075809 UP000078541 UP000192223 UP000053097 UP000279307 UP000215335 UP000009046 UP000002358 UP000007266 UP000015103 UP000053105 UP000242188 UP000069940 UP000249989 UP000008820 UP000245119 UP000030742 UP000092462 UP000076407 UP000075902 UP000075840 UP000075903 UP000002320 UP000000673 UP000030765 UP000076408 UP000075885 UP000030746 UP000019118 UP000075880 UP000075920 UP000092461 UP000001555 UP000075883 UP000076420 UP000075900 UP000000305 UP000015101 UP000075881 UP000215902 UP000095301 UP000183832 UP000075882 UP000069272 UP000037069 UP000014760
UP000027135 UP000076502 UP000235965 UP000008237 UP000036403 UP000078542 UP000078540 UP000005205 UP000078492 UP000007755 UP000242457 UP000005203 UP000075809 UP000078541 UP000192223 UP000053097 UP000279307 UP000215335 UP000009046 UP000002358 UP000007266 UP000015103 UP000053105 UP000242188 UP000069940 UP000249989 UP000008820 UP000245119 UP000030742 UP000092462 UP000076407 UP000075902 UP000075840 UP000075903 UP000002320 UP000000673 UP000030765 UP000076408 UP000075885 UP000030746 UP000019118 UP000075880 UP000075920 UP000092461 UP000001555 UP000075883 UP000076420 UP000075900 UP000000305 UP000015101 UP000075881 UP000215902 UP000095301 UP000183832 UP000075882 UP000069272 UP000037069 UP000014760
Interpro
ProteinModelPortal
H9J5C1
A0A2A4IVP2
A0A194R1Z9
A0A194Q3L6
A0A3S2NZ56
S4NSQ2
+ More
A0A212FAF2 A0A2H1WCW8 A0A067QQE5 A0A154PHZ2 A0A2J7QCA4 E9IDT2 E2B6Y7 A0A0J7NJC2 A0A195C2V5 A0A195BB51 A0A158NN53 A0A151IWH2 F4W8C7 A0A2A3E1P5 A0A087ZSJ5 A0A310SK01 A0A151X288 A0A195FAZ1 A0A1W4X1Z7 A0A1B6GJV2 A0A026WFT3 A0A1B6LL71 A0A1Y1LPU7 A0A232ESW5 A0A1B6HMU1 E0VMB1 K7J661 D6X0K0 A0A0C9QC64 A0A0N7Z9B2 R4FNT4 A0A0M8ZT85 A0A0V0G8C8 A0A069DU69 A0A210PMW5 A0A293MX40 A0A2R5LHA1 A0A1Z5LAI6 A0A182HCN0 U5EXK8 Q0IFL2 A0A2T7NVP9 T1IJL2 U4UF38 A0A1Q3F4B3 A0A1B0D5N8 A0A182XKF9 A0A182UB01 A0A2C9GQH2 A0A182VAW2 B0W8I1 W8CA62 W5JVX2 A0A0B7B8P4 L7MGZ0 A0A023FXK4 A0A084VTP9 A0A182XXI8 A0A131XCH3 A0A224Z0K8 A0A131Z1S1 A0A182PBG2 V4AYE4 A0A2R7X2A1 N6U3Y5 A0A023GNL1 G3MH90 A0A182IUZ6 A0A3F2Z113 A0A1B0CJ20 B7PYR8 A0A182MWY7 A0A2C9JD95 A0A1Y9HER1 A0A2M4ANB7 E9H9P0 A0A034WBU7 A0A224XQ29 A0A023FK87 T1FSY2 A0A131XXE0 A0A0K8W5T8 A0A182K3C8 A0A0P5XRN9 A0A267FV70 A0A1I8N241 A0A1J1HUP3 A0A182LCW5 A0A182FAX4 K4G9W9 A0A0T6B5H1 A0A0L0CDX9 A0A267F687 R7V1X6
A0A212FAF2 A0A2H1WCW8 A0A067QQE5 A0A154PHZ2 A0A2J7QCA4 E9IDT2 E2B6Y7 A0A0J7NJC2 A0A195C2V5 A0A195BB51 A0A158NN53 A0A151IWH2 F4W8C7 A0A2A3E1P5 A0A087ZSJ5 A0A310SK01 A0A151X288 A0A195FAZ1 A0A1W4X1Z7 A0A1B6GJV2 A0A026WFT3 A0A1B6LL71 A0A1Y1LPU7 A0A232ESW5 A0A1B6HMU1 E0VMB1 K7J661 D6X0K0 A0A0C9QC64 A0A0N7Z9B2 R4FNT4 A0A0M8ZT85 A0A0V0G8C8 A0A069DU69 A0A210PMW5 A0A293MX40 A0A2R5LHA1 A0A1Z5LAI6 A0A182HCN0 U5EXK8 Q0IFL2 A0A2T7NVP9 T1IJL2 U4UF38 A0A1Q3F4B3 A0A1B0D5N8 A0A182XKF9 A0A182UB01 A0A2C9GQH2 A0A182VAW2 B0W8I1 W8CA62 W5JVX2 A0A0B7B8P4 L7MGZ0 A0A023FXK4 A0A084VTP9 A0A182XXI8 A0A131XCH3 A0A224Z0K8 A0A131Z1S1 A0A182PBG2 V4AYE4 A0A2R7X2A1 N6U3Y5 A0A023GNL1 G3MH90 A0A182IUZ6 A0A3F2Z113 A0A1B0CJ20 B7PYR8 A0A182MWY7 A0A2C9JD95 A0A1Y9HER1 A0A2M4ANB7 E9H9P0 A0A034WBU7 A0A224XQ29 A0A023FK87 T1FSY2 A0A131XXE0 A0A0K8W5T8 A0A182K3C8 A0A0P5XRN9 A0A267FV70 A0A1I8N241 A0A1J1HUP3 A0A182LCW5 A0A182FAX4 K4G9W9 A0A0T6B5H1 A0A0L0CDX9 A0A267F687 R7V1X6
PDB
6QI9
E-value=0,
Score=1798
Ontologies
GO
GO:0031011
GO:0006325
GO:0005524
GO:0006281
GO:0035267
GO:0097255
GO:0043141
GO:0005634
GO:0004386
GO:0006338
GO:0006357
GO:0000492
GO:0016573
GO:0004003
GO:0000812
GO:0042127
GO:0006310
GO:0003713
GO:0051301
GO:0007049
GO:0090263
GO:0016021
GO:0016491
GO:0003678
GO:0006468
GO:0020037
GO:0006397
GO:0005737
GO:0048869
GO:0003676
GO:0015930
PANTHER
Topology
Subcellular location
Nucleus
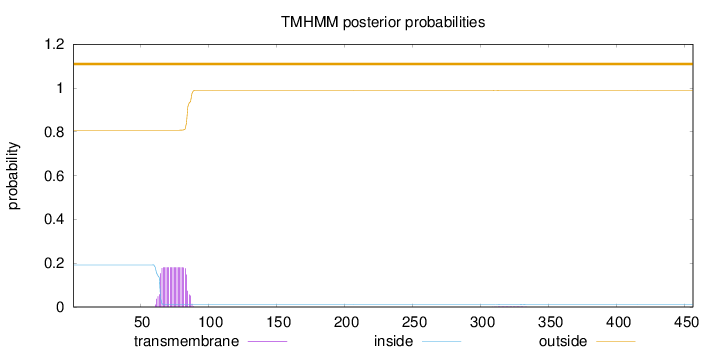
Length:
456
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.95908
Exp number, first 60 AAs:
0.00118
Total prob of N-in:
0.19222
outside
1 - 456
Population Genetic Test Statistics
Pi
395.630188
Theta
213.628458
Tajima's D
1.794564
CLR
0.421185
CSRT
0.844757762111894
Interpretation
Uncertain
