Gene
KWMTBOMO13285
Pre Gene Modal
BGIBMGA004712
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_treacle_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.287
Sequence
CDS
ATGGTAGTGGGCGAAAATAAGCGCACCGCCAATATCATGGACGGGATCGAAAACACCGGCGACGTGCGTATGCACGACCACCGGCTGAGGCTCAAACAAAGATTTGACATAGTAAGAAAATTAGGACAAGGCACGTACGGCAAGGTGCAGCTCGGTATCAACAAGAAGACCGGTCAGGAAGTGGCCATTAAGACAATAAAGAAATGCAAGATCGAATCTGAAGCGGATCTGATTCGTATAAGAAGAGAAGTACAGATCATGTCGTCGGTCAGACATCCAAACATTGTACATATCTATGAAGTATTCGAAAACAGTGAAAAAATGATCCTAGTGATGGAATACTGTTCCGGTGGCGAACTGTACGATTATCTCAGTCAGAGGAAAGTGTTACAAGAGGATGAAGCCCGAAGGCTTTTCAGACAAATCGCCACTGCGGTGTATTACTGTCACATTCACAAAATCTGTCACAGAGATTTGAAATTAGAAAACGTATTGTTAGACGATACTGGAAGCGCTAAGATCGCGGACTTTGGTTTGTCGAACGTTTTCAAAGAAACGTCTCTGCTGTCCACGTTCTGCGGGTCACCTCTGTACGCGTCTCCTGAAATCGTCAAAGGAACACCTTATATCGGGCCCGAGGTGGATTGCTGGTCTCTAGGCGTTCTCCTGTACACTCTAGTCTACGGAGCCATGCCGTTCGATGGCTCCAACTTCAAGCGGCTGGTCAGGCAAATCAGTAACGGAGACTATTACGAGCCCAGGAATCCTTCGAGTGCCTCGCCCTTAATCCGTGATATGCTCACGGTCGATCCTTTGAAACGTGCTGACATCGCCTACATATGTGATCATCCGTGGGTGAACACTGGCTGCGAGACGTCATGCCTGGAGATGGCTGAAGCGCTAGCCGCCGAAACTCCAGTGCGTTTGGATCTGCTGCTCTCGCTGGCTCCGGCTGCTCCTTCTAACTCCAACTCTGTTATGGTTCCGGCTGAGGCCGAAATGGGTCCGGAAGAGAGCTGCACCGACCTGACAAGCTCGACGTCGATGGCCCTAGAGCCCAGCAATTCCGCTGAAAAGAGAATCCTTGAATTGGTTGCAGAGGGCGGTGAAGACGCAATAAAACCATCGCCGACGAGAACAATAGTTTCTGCAGGGGAGAACAAACGGAAATTGGAGCTCGCCATGTCTGCTAGTGGACTACAGCGGAAGAAAGAGAGAGTGCCGAGCGCGGCAAGTATCGCGCCCCCCGGCGGCTCGGTGCCGGAGGAGAGCCCCGAGGCGCAGGACGCGGCCACCACGCCAACACCCCCCGACGAACCACCCCCACTCCTCGCTGACGAGTCGGTACAGCCATCGCTGAAGTCTTCGGCCACCATGACGATAACGCCCACACCAGTAGAGCTCGTAAAAGACCTGACCAAGGAGATCATCAGGGACGCTCCGAAAGACGAAGAGAAGGACAAGTCTGGGGCGAGGCCACGGGTCGTCATCAAGAAAAAACCTTCACCGACTAAAACGGTTGAAGAGAAACCCCCGCCGCCTGACACTGTTGAAGAGCTCAAGTTAGCGGAACAAGAGGAGCCGAAGTCAAACGCAGTAGCGTCGGCGGCCGACAAACTCACTTCTCTGAGCATCGCGCAGCAACCTGCGAACCAAGCTCCCGCTAAACCTAAGAAACTCTCAATACCAGGTGGAAATGTCGGTAACTTCAAGGAACAATTCGAGAAACGGGCGAATTCTGTATCAAAGCCAGTGGTATCGAAGATCGCCAAGCCCAAAGCTCAAAGTGAAGACCGCGAGCTTTCGAACAAAACTTCAGACAATGGCACTGTTCGACTGCAACAGATCGGAATTGAACCTGCACCGTCTCCGATCGAGCTGGAAATGAATCGGGGCGGCGTGAAGAAAACCGAGAGCGAACCGACACACGGCGCGGCCGCGGTCGACCCCTCCGTGCTCCTCAGCGACGCTAGACGAAGTCTCCAGAATTCCATGGCGAAGCTTGTGGAAGAGAAAACAGGAGAGGAGAGTCGTCGTCGGGCTGCGCGCGATATTATATCGTCCGCCATCCGCACAGGCAAGCCTCCGGTGCCTTACGGGCGCTCGTCGTCGGCGGGCGTGGCGTCGCTGGTGTCGCCGCCCGGCACCCCGCCCGCCGCCGCCGCCCCCGCGCGCCTCTTCCGCACCGAGGCGCACCACCAGGTCGAGGATCATCGCAACAGGTACTACACACGCCATACGCGGTATTTTTTTTAG
Protein
MVVGENKRTANIMDGIENTGDVRMHDHRLRLKQRFDIVRKLGQGTYGKVQLGINKKTGQEVAIKTIKKCKIESEADLIRIRREVQIMSSVRHPNIVHIYEVFENSEKMILVMEYCSGGELYDYLSQRKVLQEDEARRLFRQIATAVYYCHIHKICHRDLKLENVLLDDTGSAKIADFGLSNVFKETSLLSTFCGSPLYASPEIVKGTPYIGPEVDCWSLGVLLYTLVYGAMPFDGSNFKRLVRQISNGDYYEPRNPSSASPLIRDMLTVDPLKRADIAYICDHPWVNTGCETSCLEMAEALAAETPVRLDLLLSLAPAAPSNSNSVMVPAEAEMGPEESCTDLTSSTSMALEPSNSAEKRILELVAEGGEDAIKPSPTRTIVSAGENKRKLELAMSASGLQRKKERVPSAASIAPPGGSVPEESPEAQDAATTPTPPDEPPPLLADESVQPSLKSSATMTITPTPVELVKDLTKEIIRDAPKDEEKDKSGARPRVVIKKKPSPTKTVEEKPPPPDTVEELKLAEQEEPKSNAVASAADKLTSLSIAQQPANQAPAKPKKLSIPGGNVGNFKEQFEKRANSVSKPVVSKIAKPKAQSEDRELSNKTSDNGTVRLQQIGIEPAPSPIELEMNRGGVKKTESEPTHGAAAVDPSVLLSDARRSLQNSMAKLVEEKTGEESRRRAARDIISSAIRTGKPPVPYGRSSSAGVASLVSPPGTPPAAAAPARLFRTEAHHQVEDHRNRYYTRHTRYFF
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
PDB
6E4W
E-value=3.85533e-66,
Score=641
Ontologies
GO
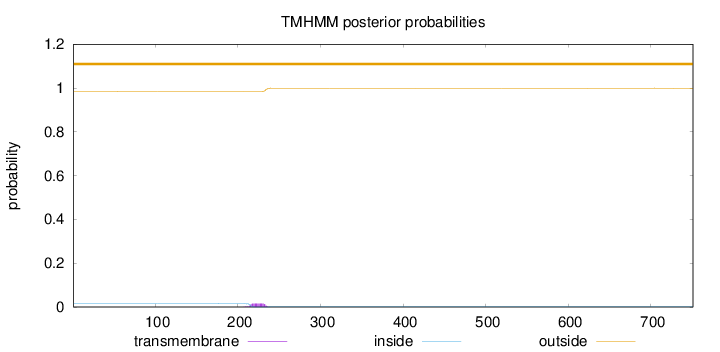
Topology
Length:
751
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31246
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01637
outside
1 - 751
Population Genetic Test Statistics
Pi
253.308339
Theta
177.802505
Tajima's D
1.265802
CLR
0.438606
CSRT
0.729963501824909
Interpretation
Uncertain
