Gene
KWMTBOMO13279
Pre Gene Modal
BGIBMGA004717
Annotation
PREDICTED:_cytochrome_P450_4c21-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.047 Mitochondrial Reliability : 1.308
Sequence
CDS
ATGTTTGTAAGTGGCTATACATCCGGTAACGTTTTGCCTGCCGGTTCAGGGATAATGGTTTCTATTTGGGGAATTCACAGAGACCCAAAGTACTGGGGTCCGGAGGCGGAACACTTCGATCCAGACAGGTTTCTGCCGGAGCGTTTTAATCTCGAGCACCCATGCTGCTACATGCCGTTCAGTAGCGGCCCTAGGAACTGTCTGGGTTATCAGTACGCAATGATATCGATTAAAACTTCTTTATCTGCAATACTGCGGAGATACAAGGTAGTAGGTGAACCGGAGAAAGGTCCTGTTCCTCAAATCAGGGTCAAACTGGATATCATGATGAAGTCAGTGAACGGATGCCAGGTGGCTTTGGAGAGAAGGCCAACAAAATAG
Protein
MFVSGYTSGNVLPAGSGIMVSIWGIHRDPKYWGPEAEHFDPDRFLPERFNLEHPCCYMPFSSGPRNCLGYQYAMISIKTSLSAILRRYKVVGEPEKGPVPQIRVKLDIMMKSVNGCQVALERRPTK
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9J5C7
H9J5D0
H9J5C8
H9J5C6
H9J5C4
A0A0L7KZU6
+ More
A0A0L7KQH7 A0A0L7L4L4 A0A0L7L583 A0A194Q3L1 A0A194R0G6 A0A2A4IYA1 A0A0K8TUH6 A0A194R208 A0A248QEI3 S4VGT4 J7FIT6 A0A212FEU5 A0A1V0D9H7 A0A194PZ23 A0A194R193 A0A3B0E7F1 A0A0C5C579 Q4R1I7 A0A212EGV4 A0A194PY94 A0A194R5Z5 A0A1V0D9H9 X5DB16 A0A3S2TR28 A0A0L7K431 A0A1V0D9I5 A0A3S2M7I8 A0A248QED4 X5DB21 A0A0L7KSI3 A0A3S2PCK4 A0A2H1VP75 A0A2A4JHV7 A0A068EU38 A0A0K8TUI4 A0A2A4IT41 A0A0L7KP65 A0A2A4JGQ9 A0A2A4JGE5 A0A1E1WU74 A0A194REI2 A0A286QUH1 A0A068EVJ0 A0A182H5R9 A0A2H1WUL9 D2A4X1 Q16P40 A0A194PZG6 B0XDU2 Q1DGU5 A0A3Q0IPK1 B0XDU1 B0X9M1 B0XDU3 A0A182PYY7 A0A1L8E4G7 A0A182G682 A0A139WGR2 Q16P39 A0A182QJY3 A0A194PYM9 A0A0D6A1C6 A0A2H1WUI5 A0A1W6L1D9 A0A182FAN8 A0A182S8E5 Q16L49 A0A2H1VMK4 A0A2Z5SAV1 A0A182J9H5 A0A182TYW8 E2BXW3 B0X9M4 A0A182Q269 A0A2A4JY96 A0A068EU45 A0A3F2Z121 A0A182H5S0 W8P916 Q2PZV9 A0A0K8TV31 A0A0C9RFM1 Q16L52 A0A139WKY0 A0A1S4FX07 Q16L55 A0A2J7QZN6 A0A084WP81 A0A182TVR0 A0A0K8TUL8 D6X3W8
A0A0L7KQH7 A0A0L7L4L4 A0A0L7L583 A0A194Q3L1 A0A194R0G6 A0A2A4IYA1 A0A0K8TUH6 A0A194R208 A0A248QEI3 S4VGT4 J7FIT6 A0A212FEU5 A0A1V0D9H7 A0A194PZ23 A0A194R193 A0A3B0E7F1 A0A0C5C579 Q4R1I7 A0A212EGV4 A0A194PY94 A0A194R5Z5 A0A1V0D9H9 X5DB16 A0A3S2TR28 A0A0L7K431 A0A1V0D9I5 A0A3S2M7I8 A0A248QED4 X5DB21 A0A0L7KSI3 A0A3S2PCK4 A0A2H1VP75 A0A2A4JHV7 A0A068EU38 A0A0K8TUI4 A0A2A4IT41 A0A0L7KP65 A0A2A4JGQ9 A0A2A4JGE5 A0A1E1WU74 A0A194REI2 A0A286QUH1 A0A068EVJ0 A0A182H5R9 A0A2H1WUL9 D2A4X1 Q16P40 A0A194PZG6 B0XDU2 Q1DGU5 A0A3Q0IPK1 B0XDU1 B0X9M1 B0XDU3 A0A182PYY7 A0A1L8E4G7 A0A182G682 A0A139WGR2 Q16P39 A0A182QJY3 A0A194PYM9 A0A0D6A1C6 A0A2H1WUI5 A0A1W6L1D9 A0A182FAN8 A0A182S8E5 Q16L49 A0A2H1VMK4 A0A2Z5SAV1 A0A182J9H5 A0A182TYW8 E2BXW3 B0X9M4 A0A182Q269 A0A2A4JY96 A0A068EU45 A0A3F2Z121 A0A182H5S0 W8P916 Q2PZV9 A0A0K8TV31 A0A0C9RFM1 Q16L52 A0A139WKY0 A0A1S4FX07 Q16L55 A0A2J7QZN6 A0A084WP81 A0A182TVR0 A0A0K8TUL8 D6X3W8
Pubmed
EMBL
BABH01037061
BABH01037064
BABH01037065
BABH01037058
BABH01037059
BABH01037052
+ More
JTDY01004128 KOB68554.1 JTDY01007290 KOB65321.1 JTDY01002945 KOB70438.1 KOB70439.1 KQ459586 KPI98000.1 KQ460883 KPJ11283.1 NWSH01005164 PCG64388.1 GCVX01000086 JAI18144.1 KPJ11285.1 KX443470 ASO98045.1 KC789756 AGO62011.1 JX310094 AFP20605.1 AGBW02008898 OWR52238.1 KY212088 ARA91652.1 KPI97999.1 KPJ11284.1 RBVL01000320 RKO09374.1 KP001145 AJN91190.1 AB201381 BAD99563.1 AGBW02015005 OWR40717.1 KPI97998.1 KPJ11286.1 KY212090 ARA91654.1 KF701170 AHW57340.1 RSAL01000018 RVE52820.1 JTDY01011574 KOB55455.1 KY212089 ARA91653.1 RVE52821.1 KX425016 ASN63843.1 KF701175 AHW57345.1 JTDY01006414 KOB66031.1 RSAL01000100 RVE47555.1 ODYU01003376 SOQ42054.1 NWSH01001465 PCG71164.1 KM016717 AID54869.1 GCVX01000088 JAI18142.1 NWSH01007729 PCG62839.1 JTDY01007865 KOB64900.1 PCG71161.1 PCG71165.1 GDQN01000461 JAT90593.1 KQ460323 KPJ15874.1 KX443471 ASO98046.1 KM016716 AID54868.1 JXUM01004862 KQ560183 KXJ83957.1 ODYU01011123 SOQ56666.1 KQ971344 EFA05708.1 CH477795 EAT36132.2 KPI98134.1 DS232782 EDS45649.1 CH900366 EAT32374.1 EDS45648.1 DS232542 EDS43223.1 EDS45650.1 GFDF01000662 JAV13422.1 JXUM01007647 KQ560244 KXJ83621.1 KYB27142.1 EAT36133.1 AXCN02001559 KPI98133.1 AB795798 BAQ56325.1 SOQ56667.1 KX609528 ARN17935.1 CH477923 EAT35039.1 SOQ42053.1 LC326250 BBA58211.1 AXCP01008626 GL451328 EFN79463.1 EDS43226.1 NWSH01000444 PCG76362.1 KM016722 AID54874.1 JXUM01004863 JXUM01004864 JXUM01004865 KXJ83958.1 KF979121 AHL26982.1 DQ279461 ABB86762.2 GCVX01000087 JAI18143.1 GBYB01007184 GBYB01007185 JAG76951.1 JAG76952.1 EAT35036.2 KQ971326 KYB28487.1 EAT35033.2 NEVH01009068 PNF34049.1 ATLV01024976 KE525366 KFB52025.1 GCVX01000053 JAI18177.1 KQ971379 EEZ97716.1
JTDY01004128 KOB68554.1 JTDY01007290 KOB65321.1 JTDY01002945 KOB70438.1 KOB70439.1 KQ459586 KPI98000.1 KQ460883 KPJ11283.1 NWSH01005164 PCG64388.1 GCVX01000086 JAI18144.1 KPJ11285.1 KX443470 ASO98045.1 KC789756 AGO62011.1 JX310094 AFP20605.1 AGBW02008898 OWR52238.1 KY212088 ARA91652.1 KPI97999.1 KPJ11284.1 RBVL01000320 RKO09374.1 KP001145 AJN91190.1 AB201381 BAD99563.1 AGBW02015005 OWR40717.1 KPI97998.1 KPJ11286.1 KY212090 ARA91654.1 KF701170 AHW57340.1 RSAL01000018 RVE52820.1 JTDY01011574 KOB55455.1 KY212089 ARA91653.1 RVE52821.1 KX425016 ASN63843.1 KF701175 AHW57345.1 JTDY01006414 KOB66031.1 RSAL01000100 RVE47555.1 ODYU01003376 SOQ42054.1 NWSH01001465 PCG71164.1 KM016717 AID54869.1 GCVX01000088 JAI18142.1 NWSH01007729 PCG62839.1 JTDY01007865 KOB64900.1 PCG71161.1 PCG71165.1 GDQN01000461 JAT90593.1 KQ460323 KPJ15874.1 KX443471 ASO98046.1 KM016716 AID54868.1 JXUM01004862 KQ560183 KXJ83957.1 ODYU01011123 SOQ56666.1 KQ971344 EFA05708.1 CH477795 EAT36132.2 KPI98134.1 DS232782 EDS45649.1 CH900366 EAT32374.1 EDS45648.1 DS232542 EDS43223.1 EDS45650.1 GFDF01000662 JAV13422.1 JXUM01007647 KQ560244 KXJ83621.1 KYB27142.1 EAT36133.1 AXCN02001559 KPI98133.1 AB795798 BAQ56325.1 SOQ56667.1 KX609528 ARN17935.1 CH477923 EAT35039.1 SOQ42053.1 LC326250 BBA58211.1 AXCP01008626 GL451328 EFN79463.1 EDS43226.1 NWSH01000444 PCG76362.1 KM016722 AID54874.1 JXUM01004863 JXUM01004864 JXUM01004865 KXJ83958.1 KF979121 AHL26982.1 DQ279461 ABB86762.2 GCVX01000087 JAI18143.1 GBYB01007184 GBYB01007185 JAG76951.1 JAG76952.1 EAT35036.2 KQ971326 KYB28487.1 EAT35033.2 NEVH01009068 PNF34049.1 ATLV01024976 KE525366 KFB52025.1 GCVX01000053 JAI18177.1 KQ971379 EEZ97716.1
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9J5C7
H9J5D0
H9J5C8
H9J5C6
H9J5C4
A0A0L7KZU6
+ More
A0A0L7KQH7 A0A0L7L4L4 A0A0L7L583 A0A194Q3L1 A0A194R0G6 A0A2A4IYA1 A0A0K8TUH6 A0A194R208 A0A248QEI3 S4VGT4 J7FIT6 A0A212FEU5 A0A1V0D9H7 A0A194PZ23 A0A194R193 A0A3B0E7F1 A0A0C5C579 Q4R1I7 A0A212EGV4 A0A194PY94 A0A194R5Z5 A0A1V0D9H9 X5DB16 A0A3S2TR28 A0A0L7K431 A0A1V0D9I5 A0A3S2M7I8 A0A248QED4 X5DB21 A0A0L7KSI3 A0A3S2PCK4 A0A2H1VP75 A0A2A4JHV7 A0A068EU38 A0A0K8TUI4 A0A2A4IT41 A0A0L7KP65 A0A2A4JGQ9 A0A2A4JGE5 A0A1E1WU74 A0A194REI2 A0A286QUH1 A0A068EVJ0 A0A182H5R9 A0A2H1WUL9 D2A4X1 Q16P40 A0A194PZG6 B0XDU2 Q1DGU5 A0A3Q0IPK1 B0XDU1 B0X9M1 B0XDU3 A0A182PYY7 A0A1L8E4G7 A0A182G682 A0A139WGR2 Q16P39 A0A182QJY3 A0A194PYM9 A0A0D6A1C6 A0A2H1WUI5 A0A1W6L1D9 A0A182FAN8 A0A182S8E5 Q16L49 A0A2H1VMK4 A0A2Z5SAV1 A0A182J9H5 A0A182TYW8 E2BXW3 B0X9M4 A0A182Q269 A0A2A4JY96 A0A068EU45 A0A3F2Z121 A0A182H5S0 W8P916 Q2PZV9 A0A0K8TV31 A0A0C9RFM1 Q16L52 A0A139WKY0 A0A1S4FX07 Q16L55 A0A2J7QZN6 A0A084WP81 A0A182TVR0 A0A0K8TUL8 D6X3W8
A0A0L7KQH7 A0A0L7L4L4 A0A0L7L583 A0A194Q3L1 A0A194R0G6 A0A2A4IYA1 A0A0K8TUH6 A0A194R208 A0A248QEI3 S4VGT4 J7FIT6 A0A212FEU5 A0A1V0D9H7 A0A194PZ23 A0A194R193 A0A3B0E7F1 A0A0C5C579 Q4R1I7 A0A212EGV4 A0A194PY94 A0A194R5Z5 A0A1V0D9H9 X5DB16 A0A3S2TR28 A0A0L7K431 A0A1V0D9I5 A0A3S2M7I8 A0A248QED4 X5DB21 A0A0L7KSI3 A0A3S2PCK4 A0A2H1VP75 A0A2A4JHV7 A0A068EU38 A0A0K8TUI4 A0A2A4IT41 A0A0L7KP65 A0A2A4JGQ9 A0A2A4JGE5 A0A1E1WU74 A0A194REI2 A0A286QUH1 A0A068EVJ0 A0A182H5R9 A0A2H1WUL9 D2A4X1 Q16P40 A0A194PZG6 B0XDU2 Q1DGU5 A0A3Q0IPK1 B0XDU1 B0X9M1 B0XDU3 A0A182PYY7 A0A1L8E4G7 A0A182G682 A0A139WGR2 Q16P39 A0A182QJY3 A0A194PYM9 A0A0D6A1C6 A0A2H1WUI5 A0A1W6L1D9 A0A182FAN8 A0A182S8E5 Q16L49 A0A2H1VMK4 A0A2Z5SAV1 A0A182J9H5 A0A182TYW8 E2BXW3 B0X9M4 A0A182Q269 A0A2A4JY96 A0A068EU45 A0A3F2Z121 A0A182H5S0 W8P916 Q2PZV9 A0A0K8TV31 A0A0C9RFM1 Q16L52 A0A139WKY0 A0A1S4FX07 Q16L55 A0A2J7QZN6 A0A084WP81 A0A182TVR0 A0A0K8TUL8 D6X3W8
PDB
6C94
E-value=1.0824e-17,
Score=213
Ontologies
GO
Topology
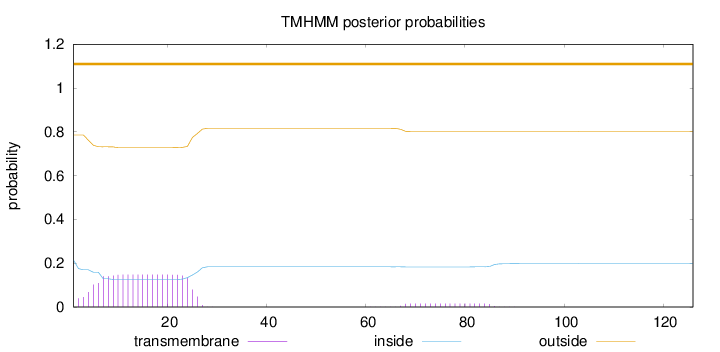
Length:
126
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.4035
Exp number, first 60 AAs:
3.11685
Total prob of N-in:
0.21467
outside
1 - 126
Population Genetic Test Statistics
Pi
37.794544
Theta
39.528911
Tajima's D
-0.400402
CLR
0
CSRT
0.267586620668967
Interpretation
Uncertain
