Gene
KWMTBOMO13278
Pre Gene Modal
BGIBMGA004721
Annotation
PREDICTED:_probable_cytochrome_P450_313a2_[Bombyx_mori]
Full name
Gustatory receptor
Location in the cell
Mitochondrial Reliability : 0.914
Sequence
CDS
ATGAAATATTTGGAGAGAGTTGTGAAGGAGTCTCTCAGATTATTCCCTCCAGTTCCGTTTATTATAAGAAAAGTTTTGGAAGATATTACGCTACCATCCGGTAACATTTTGCCTGCCGGTTCAGGAGTAGTGGTTTCTATCTGGGGAATTCACAGAGACCCAAAGTACTGGGGTCCGGAGGCGGAATACTTCGACCCAGACAGGTTTCTACCGGAGCGTTTTAATGTCGAGCACCCGTGCTGCTACATGCCGTTCAGTAGCGGTCCTAGGAATTGCCTGGGTGAGTACCGTGCTCATACCAAGATATCCACGTCGTTAGAGCCTTTGCGCTTAATCTTCTCGTCTCCACGGAACAAGAAGTTCCTTGTTGTTGTGGTATCTTGA
Protein
MKYLERVVKESLRLFPPVPFIIRKVLEDITLPSGNILPAGSGVVVSIWGIHRDPKYWGPEAEYFDPDRFLPERFNVEHPCCYMPFSSGPRNCLGEYRAHTKISTSLEPLRLIFSSPRNKKFLVVVVS
Summary
Description
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates.
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Uniprot
H9J5D1
H9J5C8
H9J5D0
H9J5C6
H9J5C7
A0A194Q3L1
+ More
S4VGT4 A0A248QEI3 A0A194R208 J7FIT6 A0A0K8TUH6 A0A194R0G6 A0A0L7K431 A0A194R193 A0A0L7L4L4 A0A194PZ23 A0A1V0D9H7 Q4R1I7 A0A2A4IYA1 A0A194R5Z5 A0A0L7K4R1 A0A194PY94 A0A0L7KZU6 X5DB16 A0A3S2TR28 A0A0L7KQH7 A0A212FEU5 A0A0C5C579 A0A3B0E7F1 A0A212EGV4 A0A0L7L530 A0A1V0D9I5 A0A1V0D9H9 X5D4I4 A0A2A4JGQ9 A0A3S2M7I8 A0A2A4JHV7 A0A2A4JGE5 A0A1E1WU74 X5DB21 A0A194REI2 A0A3S2PCK4 A0A194PZG6 A0A068EVJ0 A0A0L7KSI3 A0A0L7L583 A0A194RF00 A0A3L8DPI1 A0A2A4IT41 A0A2H1WBM8 A0A026X1C5 A0A1B6LVG5 A0A286QUH1 R4WD28 H9J588 L0N785 A0A067RD05 A0A154PPN0 A0A0D6A1C6 A0A1B0DL32 K7IXL4 A0A0A9ZHM5 A0A0A9ZFR9 A0A1B6I8B2 A0A2Z5SAV1 A0A0A9XS63 A0A2H1WUL9 B4LKW3 A0A1B6J693 A0A1B6HPU9 A0A0K8TV26 H9JHK8 A0A0K8TUI4 L0N5K2 A0A1B6JPD5 A0A194RMW0 A0A2W1C3W1 A0A0A9ZE96 A0A023F6W9 A0A182S8E5 K7J9G3 A0A1B6BYW1 A0A146MBN7 A0A336LN05 A0A1W6L1H7 Q16L52 A0A1B6HU81 A0A0A9Y5B0 A0A232FMF7 A0A0A9WLL0 A0A1E1WJF1 A0A2H1WBK5 A0A1B6H674 A0A0K8T7V9 A0A146LN63 A0A194PYM9 R4FLB1
S4VGT4 A0A248QEI3 A0A194R208 J7FIT6 A0A0K8TUH6 A0A194R0G6 A0A0L7K431 A0A194R193 A0A0L7L4L4 A0A194PZ23 A0A1V0D9H7 Q4R1I7 A0A2A4IYA1 A0A194R5Z5 A0A0L7K4R1 A0A194PY94 A0A0L7KZU6 X5DB16 A0A3S2TR28 A0A0L7KQH7 A0A212FEU5 A0A0C5C579 A0A3B0E7F1 A0A212EGV4 A0A0L7L530 A0A1V0D9I5 A0A1V0D9H9 X5D4I4 A0A2A4JGQ9 A0A3S2M7I8 A0A2A4JHV7 A0A2A4JGE5 A0A1E1WU74 X5DB21 A0A194REI2 A0A3S2PCK4 A0A194PZG6 A0A068EVJ0 A0A0L7KSI3 A0A0L7L583 A0A194RF00 A0A3L8DPI1 A0A2A4IT41 A0A2H1WBM8 A0A026X1C5 A0A1B6LVG5 A0A286QUH1 R4WD28 H9J588 L0N785 A0A067RD05 A0A154PPN0 A0A0D6A1C6 A0A1B0DL32 K7IXL4 A0A0A9ZHM5 A0A0A9ZFR9 A0A1B6I8B2 A0A2Z5SAV1 A0A0A9XS63 A0A2H1WUL9 B4LKW3 A0A1B6J693 A0A1B6HPU9 A0A0K8TV26 H9JHK8 A0A0K8TUI4 L0N5K2 A0A1B6JPD5 A0A194RMW0 A0A2W1C3W1 A0A0A9ZE96 A0A023F6W9 A0A182S8E5 K7J9G3 A0A1B6BYW1 A0A146MBN7 A0A336LN05 A0A1W6L1H7 Q16L52 A0A1B6HU81 A0A0A9Y5B0 A0A232FMF7 A0A0A9WLL0 A0A1E1WJF1 A0A2H1WBK5 A0A1B6H674 A0A0K8T7V9 A0A146LN63 A0A194PYM9 R4FLB1
Pubmed
EMBL
BABH01037067
BABH01037068
BABH01037061
BABH01037064
BABH01037065
BABH01037058
+ More
BABH01037059 KQ459586 KPI98000.1 KC789756 AGO62011.1 KX443470 ASO98045.1 KQ460883 KPJ11285.1 JX310094 AFP20605.1 GCVX01000086 JAI18144.1 KPJ11283.1 JTDY01011574 KOB55455.1 KPJ11284.1 JTDY01002945 KOB70438.1 KPI97999.1 KY212088 ARA91652.1 AB201381 BAD99563.1 NWSH01005164 PCG64388.1 KPJ11286.1 JTDY01010731 KOB58062.1 KPI97998.1 JTDY01004128 KOB68554.1 KF701170 AHW57340.1 RSAL01000018 RVE52820.1 JTDY01007290 KOB65321.1 AGBW02008898 OWR52238.1 KP001145 AJN91190.1 RBVL01000320 RKO09374.1 AGBW02015005 OWR40717.1 KOB70436.1 KY212089 ARA91653.1 KY212090 ARA91654.1 KF701171 AHW57341.1 NWSH01001465 PCG71161.1 RVE52821.1 PCG71164.1 PCG71165.1 GDQN01000461 JAT90593.1 KF701175 AHW57345.1 KQ460323 KPJ15874.1 RSAL01000100 RVE47555.1 KPI98134.1 KM016716 AID54868.1 JTDY01006414 KOB66031.1 KOB70439.1 KPJ15875.1 QOIP01000005 RLU22345.1 NWSH01007729 PCG62839.1 ODYU01007567 SOQ50447.1 KK107042 EZA61816.1 GEBQ01012296 JAT27681.1 KX443471 ASO98046.1 AK417154 BAN20369.1 BABH01039691 AB436841 BAM73795.1 KK852542 KDR21746.1 KQ435012 KZC13846.1 AB795798 BAQ56325.1 AJVK01069147 AAZX01012601 GBHO01000221 JAG43383.1 GBHO01000223 GBHO01000220 JAG43381.1 JAG43384.1 GECU01024542 JAS83164.1 LC326250 BBA58211.1 GBHO01023664 GBHO01011635 JAG19940.1 JAG31969.1 ODYU01011123 SOQ56666.1 CH940648 EDW60767.2 GECU01013063 JAS94643.1 GECU01031023 JAS76683.1 GCVX01000092 JAI18138.1 BABH01012758 GCVX01000088 JAI18142.1 AK343188 BAM73878.1 GECU01006585 JAT01122.1 KQ459989 KPJ18754.1 KZ149894 PZC78853.1 GBHO01000835 JAG42769.1 GBBI01001581 JAC17131.1 GEDC01031123 JAS06175.1 GDHC01002402 JAQ16227.1 UFQT01000022 SSX17979.1 KX609525 ARN17932.1 CH477923 EAT35036.2 GECU01029511 JAS78195.1 GBHO01016783 JAG26821.1 NNAY01000026 OXU31835.1 GBHO01034980 JAG08624.1 GDQN01003911 JAT87143.1 SOQ50448.1 GECU01037548 JAS70158.1 GBRD01004571 JAG61250.1 GDHC01009820 JAQ08809.1 KPI98133.1 ACPB03008558 GAHY01001498 JAA76012.1
BABH01037059 KQ459586 KPI98000.1 KC789756 AGO62011.1 KX443470 ASO98045.1 KQ460883 KPJ11285.1 JX310094 AFP20605.1 GCVX01000086 JAI18144.1 KPJ11283.1 JTDY01011574 KOB55455.1 KPJ11284.1 JTDY01002945 KOB70438.1 KPI97999.1 KY212088 ARA91652.1 AB201381 BAD99563.1 NWSH01005164 PCG64388.1 KPJ11286.1 JTDY01010731 KOB58062.1 KPI97998.1 JTDY01004128 KOB68554.1 KF701170 AHW57340.1 RSAL01000018 RVE52820.1 JTDY01007290 KOB65321.1 AGBW02008898 OWR52238.1 KP001145 AJN91190.1 RBVL01000320 RKO09374.1 AGBW02015005 OWR40717.1 KOB70436.1 KY212089 ARA91653.1 KY212090 ARA91654.1 KF701171 AHW57341.1 NWSH01001465 PCG71161.1 RVE52821.1 PCG71164.1 PCG71165.1 GDQN01000461 JAT90593.1 KF701175 AHW57345.1 KQ460323 KPJ15874.1 RSAL01000100 RVE47555.1 KPI98134.1 KM016716 AID54868.1 JTDY01006414 KOB66031.1 KOB70439.1 KPJ15875.1 QOIP01000005 RLU22345.1 NWSH01007729 PCG62839.1 ODYU01007567 SOQ50447.1 KK107042 EZA61816.1 GEBQ01012296 JAT27681.1 KX443471 ASO98046.1 AK417154 BAN20369.1 BABH01039691 AB436841 BAM73795.1 KK852542 KDR21746.1 KQ435012 KZC13846.1 AB795798 BAQ56325.1 AJVK01069147 AAZX01012601 GBHO01000221 JAG43383.1 GBHO01000223 GBHO01000220 JAG43381.1 JAG43384.1 GECU01024542 JAS83164.1 LC326250 BBA58211.1 GBHO01023664 GBHO01011635 JAG19940.1 JAG31969.1 ODYU01011123 SOQ56666.1 CH940648 EDW60767.2 GECU01013063 JAS94643.1 GECU01031023 JAS76683.1 GCVX01000092 JAI18138.1 BABH01012758 GCVX01000088 JAI18142.1 AK343188 BAM73878.1 GECU01006585 JAT01122.1 KQ459989 KPJ18754.1 KZ149894 PZC78853.1 GBHO01000835 JAG42769.1 GBBI01001581 JAC17131.1 GEDC01031123 JAS06175.1 GDHC01002402 JAQ16227.1 UFQT01000022 SSX17979.1 KX609525 ARN17932.1 CH477923 EAT35036.2 GECU01029511 JAS78195.1 GBHO01016783 JAG26821.1 NNAY01000026 OXU31835.1 GBHO01034980 JAG08624.1 GDQN01003911 JAT87143.1 SOQ50448.1 GECU01037548 JAS70158.1 GBRD01004571 JAG61250.1 GDHC01009820 JAQ08809.1 KPI98133.1 ACPB03008558 GAHY01001498 JAA76012.1
Proteomes
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9J5D1
H9J5C8
H9J5D0
H9J5C6
H9J5C7
A0A194Q3L1
+ More
S4VGT4 A0A248QEI3 A0A194R208 J7FIT6 A0A0K8TUH6 A0A194R0G6 A0A0L7K431 A0A194R193 A0A0L7L4L4 A0A194PZ23 A0A1V0D9H7 Q4R1I7 A0A2A4IYA1 A0A194R5Z5 A0A0L7K4R1 A0A194PY94 A0A0L7KZU6 X5DB16 A0A3S2TR28 A0A0L7KQH7 A0A212FEU5 A0A0C5C579 A0A3B0E7F1 A0A212EGV4 A0A0L7L530 A0A1V0D9I5 A0A1V0D9H9 X5D4I4 A0A2A4JGQ9 A0A3S2M7I8 A0A2A4JHV7 A0A2A4JGE5 A0A1E1WU74 X5DB21 A0A194REI2 A0A3S2PCK4 A0A194PZG6 A0A068EVJ0 A0A0L7KSI3 A0A0L7L583 A0A194RF00 A0A3L8DPI1 A0A2A4IT41 A0A2H1WBM8 A0A026X1C5 A0A1B6LVG5 A0A286QUH1 R4WD28 H9J588 L0N785 A0A067RD05 A0A154PPN0 A0A0D6A1C6 A0A1B0DL32 K7IXL4 A0A0A9ZHM5 A0A0A9ZFR9 A0A1B6I8B2 A0A2Z5SAV1 A0A0A9XS63 A0A2H1WUL9 B4LKW3 A0A1B6J693 A0A1B6HPU9 A0A0K8TV26 H9JHK8 A0A0K8TUI4 L0N5K2 A0A1B6JPD5 A0A194RMW0 A0A2W1C3W1 A0A0A9ZE96 A0A023F6W9 A0A182S8E5 K7J9G3 A0A1B6BYW1 A0A146MBN7 A0A336LN05 A0A1W6L1H7 Q16L52 A0A1B6HU81 A0A0A9Y5B0 A0A232FMF7 A0A0A9WLL0 A0A1E1WJF1 A0A2H1WBK5 A0A1B6H674 A0A0K8T7V9 A0A146LN63 A0A194PYM9 R4FLB1
S4VGT4 A0A248QEI3 A0A194R208 J7FIT6 A0A0K8TUH6 A0A194R0G6 A0A0L7K431 A0A194R193 A0A0L7L4L4 A0A194PZ23 A0A1V0D9H7 Q4R1I7 A0A2A4IYA1 A0A194R5Z5 A0A0L7K4R1 A0A194PY94 A0A0L7KZU6 X5DB16 A0A3S2TR28 A0A0L7KQH7 A0A212FEU5 A0A0C5C579 A0A3B0E7F1 A0A212EGV4 A0A0L7L530 A0A1V0D9I5 A0A1V0D9H9 X5D4I4 A0A2A4JGQ9 A0A3S2M7I8 A0A2A4JHV7 A0A2A4JGE5 A0A1E1WU74 X5DB21 A0A194REI2 A0A3S2PCK4 A0A194PZG6 A0A068EVJ0 A0A0L7KSI3 A0A0L7L583 A0A194RF00 A0A3L8DPI1 A0A2A4IT41 A0A2H1WBM8 A0A026X1C5 A0A1B6LVG5 A0A286QUH1 R4WD28 H9J588 L0N785 A0A067RD05 A0A154PPN0 A0A0D6A1C6 A0A1B0DL32 K7IXL4 A0A0A9ZHM5 A0A0A9ZFR9 A0A1B6I8B2 A0A2Z5SAV1 A0A0A9XS63 A0A2H1WUL9 B4LKW3 A0A1B6J693 A0A1B6HPU9 A0A0K8TV26 H9JHK8 A0A0K8TUI4 L0N5K2 A0A1B6JPD5 A0A194RMW0 A0A2W1C3W1 A0A0A9ZE96 A0A023F6W9 A0A182S8E5 K7J9G3 A0A1B6BYW1 A0A146MBN7 A0A336LN05 A0A1W6L1H7 Q16L52 A0A1B6HU81 A0A0A9Y5B0 A0A232FMF7 A0A0A9WLL0 A0A1E1WJF1 A0A2H1WBK5 A0A1B6H674 A0A0K8T7V9 A0A146LN63 A0A194PYM9 R4FLB1
PDB
6C93
E-value=1.42698e-21,
Score=247
Ontologies
GO
Topology
Subcellular location
Cell membrane
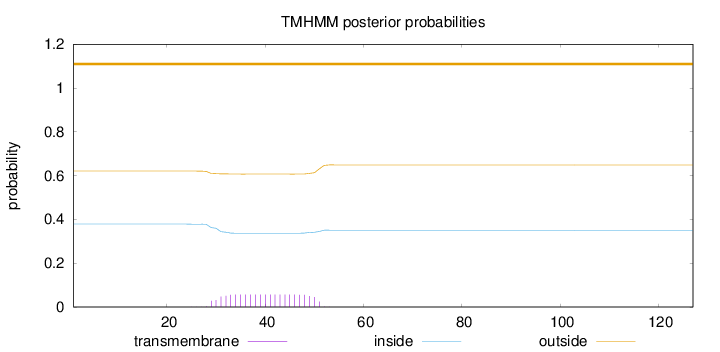
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.18338
Exp number, first 60 AAs:
1.18338
Total prob of N-in:
0.37856
outside
1 - 127
Population Genetic Test Statistics
Pi
233.005494
Theta
180.278187
Tajima's D
0.642349
CLR
0
CSRT
0.557522123893805
Interpretation
Uncertain
