Gene
KWMTBOMO13271
Annotation
olfactory_receptor_[Bombyx_mori]
Full name
Odorant receptor
+ More
Protein kinase C
Protein kinase C
Location in the cell
PlasmaMembrane Reliability : 1.482
Sequence
CDS
ATGTTTATATACTGCTATTACGGTAACGTTGTCAGCTACGAGAGCAAATATATAAATACTTCGTTGTATTTGAGTGATTGGTCATCAGCATCGCCAGGTGTCCGCAAAATGTTTCTGATCGTGATGCCGCGGTGGACGAGGCCGCTGGTAGTGCGAATCGCTAGAGTCGTACCCCTATCTTTGGACAGTTTCGTTTCTATAGTCAAATCGTCATATACTTTGTATACAATATTAGTAACAAAGAACAGCATCGCGTCAGAAGAGGCAATAATGTAA
Protein
MFIYCYYGNVVSYESKYINTSLYLSDWSSASPGVRKMFLIVMPRWTRPLVVRIARVVPLSLDSFVSIVKSSYTLYTILVTKNSIASEEAIM
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. PKC subfamily.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. PKC subfamily.
Feature
chain Protein kinase C
Uniprot
C4B7X9
A0A386H9F6
A0A1B3P5P9
A0A2H1WX32
A0A345BEX4
A0A2H1W412
+ More
A0A223HDE9 A0A0K8TUQ7 A0A223HD70 C4B7Y0 A0A076E988 A0A1B3B771 A0A076E7L4 A0A2K8GKS8 A0A1Y9TJU5 H9A5P9 A0A1X9PBW7 A0A1B3P5S2 A0A0B4ZYD8 A0A0S1TQQ0 A0A2H1VC39 A0A386H9E8 A0A146JVL7 A0A0E4B5J3 A0A2L1TG09 A0A2A4J3V9 A0A075T2X6 M1R8K1 A0A1B3B782 A0A2H1VDP1 A0A1B3B786 A0A0S1TR01 A0A1B2G2N1 A0A2W1BE83 A0A2A4JAD5 A0A3Q8HD86 Q6A1J7 R4JNR7 A0A2L1TG38 R4JSG1 A0A2W1BC37 A0A194R0D6 A0A1B3B758 A0A3Q8HDN8 H9J899 A0A1S5XXP5 A0A0U3BFE1 A0A1S5XXR4 A0A075T633 A0A1S5XXM8 A0A0U3BYM9 A0A097ITX2 A0A0B5CUS8 C4B7V7 A0A0B5GEV8 A0A097IYI2 A0A146JYN2 A0A1X9PEX2 Q0EEF5 A0A0K8TU89 C4B7Y7 A0A2K8GKT4 B9VH09 A0A0E4B5H4 A0A0B5CMT2 A0A1B3P5P6 N6TQY7 A0A2L1TG83 A0A1S5XXM2 A0A0U3AKL0 A0A2K8GL44 A0A2L1TG51 A0A1B3P5N9 A0A1V1WBS7 A0A1B3P5P5 A0A223HD65 A0A1V1WBW7 A0A2H1X0G0 A0A2H4ZBA2 A0A223HDE5 A0A0S3J2Q0 A0A2P9JY54 A0A345BEV2 A0A151K3Z1 A0A386H9G3 A0A2H1V456 A0A1W4XBP7 A0A0V0J0U7 A0A076E9B0 A0A0U3C2H9 E2ANS5 A0A2A4JDS9 A0A346D496 A0A310S620 A0A221I0C9 R9PSP6 A0A310S3M7 A0A075T7Z6 A0A2A4JPI1 A0A0U3C1J6
A0A223HDE9 A0A0K8TUQ7 A0A223HD70 C4B7Y0 A0A076E988 A0A1B3B771 A0A076E7L4 A0A2K8GKS8 A0A1Y9TJU5 H9A5P9 A0A1X9PBW7 A0A1B3P5S2 A0A0B4ZYD8 A0A0S1TQQ0 A0A2H1VC39 A0A386H9E8 A0A146JVL7 A0A0E4B5J3 A0A2L1TG09 A0A2A4J3V9 A0A075T2X6 M1R8K1 A0A1B3B782 A0A2H1VDP1 A0A1B3B786 A0A0S1TR01 A0A1B2G2N1 A0A2W1BE83 A0A2A4JAD5 A0A3Q8HD86 Q6A1J7 R4JNR7 A0A2L1TG38 R4JSG1 A0A2W1BC37 A0A194R0D6 A0A1B3B758 A0A3Q8HDN8 H9J899 A0A1S5XXP5 A0A0U3BFE1 A0A1S5XXR4 A0A075T633 A0A1S5XXM8 A0A0U3BYM9 A0A097ITX2 A0A0B5CUS8 C4B7V7 A0A0B5GEV8 A0A097IYI2 A0A146JYN2 A0A1X9PEX2 Q0EEF5 A0A0K8TU89 C4B7Y7 A0A2K8GKT4 B9VH09 A0A0E4B5H4 A0A0B5CMT2 A0A1B3P5P6 N6TQY7 A0A2L1TG83 A0A1S5XXM2 A0A0U3AKL0 A0A2K8GL44 A0A2L1TG51 A0A1B3P5N9 A0A1V1WBS7 A0A1B3P5P5 A0A223HD65 A0A1V1WBW7 A0A2H1X0G0 A0A2H4ZBA2 A0A223HDE5 A0A0S3J2Q0 A0A2P9JY54 A0A345BEV2 A0A151K3Z1 A0A386H9G3 A0A2H1V456 A0A1W4XBP7 A0A0V0J0U7 A0A076E9B0 A0A0U3C2H9 E2ANS5 A0A2A4JDS9 A0A346D496 A0A310S620 A0A221I0C9 R9PSP6 A0A310S3M7 A0A075T7Z6 A0A2A4JPI1 A0A0U3C1J6
EC Number
2.7.11.13
Pubmed
EMBL
AB472127
BAH66345.1
MG546627
AYD42250.1
KX655996
AOG12945.1
+ More
ODYU01011659 SOQ57547.1 MG816613 AXF48798.1 ODYU01006192 SOQ47830.1 KY283747 AST36406.1 GCVX01000212 JAI18018.1 KY283670 AST36329.1 AB472128 BAH66346.1 KF487655 AII01053.1 KT588151 AOE48061.1 KF487684 AII01082.1 KX585328 ARO70221.1 KX084489 ARO76444.1 JN836699 AFC91739.1 KX084498 ARO76453.1 KX655983 AOG12932.1 KJ542677 AJD81561.1 KR935723 ALM26215.1 ODYU01001743 SOQ38356.1 MG546617 AYD42240.1 GEDO01000064 JAP88562.1 LC002733 BAR43481.1 KY707189 AVF19631.1 NWSH01003581 PCG66092.1 KF768679 AIG51873.1 JX999586 AGG08876.1 KT588143 AOE48053.1 ODYU01001987 SOQ38937.1 KT588154 AOE48064.1 KR935728 ALM26220.1 KU598910 ANZ03156.1 KZ150103 PZC73462.1 NWSH01002272 PCG68726.1 MF625579 AXY83406.1 AJ748332 CAG38118.1 JX982532 AGK90007.1 KY707194 AVF19636.1 JX982545 AGK90020.1 KZ150524 PZC70606.1 KQ460883 KPJ11253.1 KT588114 AOE48024.1 MF625577 AXY83404.1 BABH01042974 KY399288 AQQ73519.1 KP975147 ALT31666.1 KY399307 AQQ73538.1 KF768681 AIG51875.1 KY399275 AQQ73506.1 KP975148 ALT31667.1 KM597207 AIT69884.1 KM678324 AJE25897.1 AB472106 BAH66323.1 KM892380 AJF23795.1 KM655543 AIT71993.1 GEDO01000030 JAP88596.1 KX084472 ARO76427.1 AB234355 BAF31195.1 GCVX01000183 JAI18047.1 AB472136 BAH66353.1 KX585350 ARO70243.1 FJ546088 ACM18061.1 LC002715 BAR43463.1 KM678306 AJE25879.1 KX655989 AOG12938.1 APGK01024014 APGK01024015 KB740529 ENN80463.1 KY707237 AVF19679.1 KY399273 AQQ73504.1 KP975156 ALT31675.1 KY225499 ARO70511.1 KY707229 AVF19671.1 KX655976 AOG12925.1 GENK01000108 JAV45805.1 KX655988 AOG12937.1 KY283654 AST36313.1 GENK01000056 JAV45857.1 ODYU01012469 SOQ58793.1 MF975460 MG766155 AUF73036.1 AVN97825.1 KY283732 AST36391.1 KT381548 ALR72554.1 KY817039 AVH87255.1 MG816591 AXF48776.1 LKEY01022044 KYN50548.1 MG546643 AYD42266.1 ODYU01000380 SOQ35064.1 GDKB01000046 JAP38450.1 KF487686 AII01084.1 KP975149 ALT31668.1 GL441322 EFN64911.1 NWSH01001847 PCG69936.1 MG859438 AXM05266.1 GGOB01000114 MCH29510.1 KY964924 ASM47077.1 GABX01000003 JAA74516.1 GGOB01000110 MCH29506.1 KF768698 AIG51892.1 NWSH01000975 PCG73322.1 KP975157 ALT31676.1
ODYU01011659 SOQ57547.1 MG816613 AXF48798.1 ODYU01006192 SOQ47830.1 KY283747 AST36406.1 GCVX01000212 JAI18018.1 KY283670 AST36329.1 AB472128 BAH66346.1 KF487655 AII01053.1 KT588151 AOE48061.1 KF487684 AII01082.1 KX585328 ARO70221.1 KX084489 ARO76444.1 JN836699 AFC91739.1 KX084498 ARO76453.1 KX655983 AOG12932.1 KJ542677 AJD81561.1 KR935723 ALM26215.1 ODYU01001743 SOQ38356.1 MG546617 AYD42240.1 GEDO01000064 JAP88562.1 LC002733 BAR43481.1 KY707189 AVF19631.1 NWSH01003581 PCG66092.1 KF768679 AIG51873.1 JX999586 AGG08876.1 KT588143 AOE48053.1 ODYU01001987 SOQ38937.1 KT588154 AOE48064.1 KR935728 ALM26220.1 KU598910 ANZ03156.1 KZ150103 PZC73462.1 NWSH01002272 PCG68726.1 MF625579 AXY83406.1 AJ748332 CAG38118.1 JX982532 AGK90007.1 KY707194 AVF19636.1 JX982545 AGK90020.1 KZ150524 PZC70606.1 KQ460883 KPJ11253.1 KT588114 AOE48024.1 MF625577 AXY83404.1 BABH01042974 KY399288 AQQ73519.1 KP975147 ALT31666.1 KY399307 AQQ73538.1 KF768681 AIG51875.1 KY399275 AQQ73506.1 KP975148 ALT31667.1 KM597207 AIT69884.1 KM678324 AJE25897.1 AB472106 BAH66323.1 KM892380 AJF23795.1 KM655543 AIT71993.1 GEDO01000030 JAP88596.1 KX084472 ARO76427.1 AB234355 BAF31195.1 GCVX01000183 JAI18047.1 AB472136 BAH66353.1 KX585350 ARO70243.1 FJ546088 ACM18061.1 LC002715 BAR43463.1 KM678306 AJE25879.1 KX655989 AOG12938.1 APGK01024014 APGK01024015 KB740529 ENN80463.1 KY707237 AVF19679.1 KY399273 AQQ73504.1 KP975156 ALT31675.1 KY225499 ARO70511.1 KY707229 AVF19671.1 KX655976 AOG12925.1 GENK01000108 JAV45805.1 KX655988 AOG12937.1 KY283654 AST36313.1 GENK01000056 JAV45857.1 ODYU01012469 SOQ58793.1 MF975460 MG766155 AUF73036.1 AVN97825.1 KY283732 AST36391.1 KT381548 ALR72554.1 KY817039 AVH87255.1 MG816591 AXF48776.1 LKEY01022044 KYN50548.1 MG546643 AYD42266.1 ODYU01000380 SOQ35064.1 GDKB01000046 JAP38450.1 KF487686 AII01084.1 KP975149 ALT31668.1 GL441322 EFN64911.1 NWSH01001847 PCG69936.1 MG859438 AXM05266.1 GGOB01000114 MCH29510.1 KY964924 ASM47077.1 GABX01000003 JAA74516.1 GGOB01000110 MCH29506.1 KF768698 AIG51892.1 NWSH01000975 PCG73322.1 KP975157 ALT31676.1
Proteomes
Interpro
SUPFAM
SSF56112
SSF56112
CDD
ProteinModelPortal
C4B7X9
A0A386H9F6
A0A1B3P5P9
A0A2H1WX32
A0A345BEX4
A0A2H1W412
+ More
A0A223HDE9 A0A0K8TUQ7 A0A223HD70 C4B7Y0 A0A076E988 A0A1B3B771 A0A076E7L4 A0A2K8GKS8 A0A1Y9TJU5 H9A5P9 A0A1X9PBW7 A0A1B3P5S2 A0A0B4ZYD8 A0A0S1TQQ0 A0A2H1VC39 A0A386H9E8 A0A146JVL7 A0A0E4B5J3 A0A2L1TG09 A0A2A4J3V9 A0A075T2X6 M1R8K1 A0A1B3B782 A0A2H1VDP1 A0A1B3B786 A0A0S1TR01 A0A1B2G2N1 A0A2W1BE83 A0A2A4JAD5 A0A3Q8HD86 Q6A1J7 R4JNR7 A0A2L1TG38 R4JSG1 A0A2W1BC37 A0A194R0D6 A0A1B3B758 A0A3Q8HDN8 H9J899 A0A1S5XXP5 A0A0U3BFE1 A0A1S5XXR4 A0A075T633 A0A1S5XXM8 A0A0U3BYM9 A0A097ITX2 A0A0B5CUS8 C4B7V7 A0A0B5GEV8 A0A097IYI2 A0A146JYN2 A0A1X9PEX2 Q0EEF5 A0A0K8TU89 C4B7Y7 A0A2K8GKT4 B9VH09 A0A0E4B5H4 A0A0B5CMT2 A0A1B3P5P6 N6TQY7 A0A2L1TG83 A0A1S5XXM2 A0A0U3AKL0 A0A2K8GL44 A0A2L1TG51 A0A1B3P5N9 A0A1V1WBS7 A0A1B3P5P5 A0A223HD65 A0A1V1WBW7 A0A2H1X0G0 A0A2H4ZBA2 A0A223HDE5 A0A0S3J2Q0 A0A2P9JY54 A0A345BEV2 A0A151K3Z1 A0A386H9G3 A0A2H1V456 A0A1W4XBP7 A0A0V0J0U7 A0A076E9B0 A0A0U3C2H9 E2ANS5 A0A2A4JDS9 A0A346D496 A0A310S620 A0A221I0C9 R9PSP6 A0A310S3M7 A0A075T7Z6 A0A2A4JPI1 A0A0U3C1J6
A0A223HDE9 A0A0K8TUQ7 A0A223HD70 C4B7Y0 A0A076E988 A0A1B3B771 A0A076E7L4 A0A2K8GKS8 A0A1Y9TJU5 H9A5P9 A0A1X9PBW7 A0A1B3P5S2 A0A0B4ZYD8 A0A0S1TQQ0 A0A2H1VC39 A0A386H9E8 A0A146JVL7 A0A0E4B5J3 A0A2L1TG09 A0A2A4J3V9 A0A075T2X6 M1R8K1 A0A1B3B782 A0A2H1VDP1 A0A1B3B786 A0A0S1TR01 A0A1B2G2N1 A0A2W1BE83 A0A2A4JAD5 A0A3Q8HD86 Q6A1J7 R4JNR7 A0A2L1TG38 R4JSG1 A0A2W1BC37 A0A194R0D6 A0A1B3B758 A0A3Q8HDN8 H9J899 A0A1S5XXP5 A0A0U3BFE1 A0A1S5XXR4 A0A075T633 A0A1S5XXM8 A0A0U3BYM9 A0A097ITX2 A0A0B5CUS8 C4B7V7 A0A0B5GEV8 A0A097IYI2 A0A146JYN2 A0A1X9PEX2 Q0EEF5 A0A0K8TU89 C4B7Y7 A0A2K8GKT4 B9VH09 A0A0E4B5H4 A0A0B5CMT2 A0A1B3P5P6 N6TQY7 A0A2L1TG83 A0A1S5XXM2 A0A0U3AKL0 A0A2K8GL44 A0A2L1TG51 A0A1B3P5N9 A0A1V1WBS7 A0A1B3P5P5 A0A223HD65 A0A1V1WBW7 A0A2H1X0G0 A0A2H4ZBA2 A0A223HDE5 A0A0S3J2Q0 A0A2P9JY54 A0A345BEV2 A0A151K3Z1 A0A386H9G3 A0A2H1V456 A0A1W4XBP7 A0A0V0J0U7 A0A076E9B0 A0A0U3C2H9 E2ANS5 A0A2A4JDS9 A0A346D496 A0A310S620 A0A221I0C9 R9PSP6 A0A310S3M7 A0A075T7Z6 A0A2A4JPI1 A0A0U3C1J6
PDB
6C70
E-value=0.00326959,
Score=89
Ontologies
KEGG
GO
PANTHER
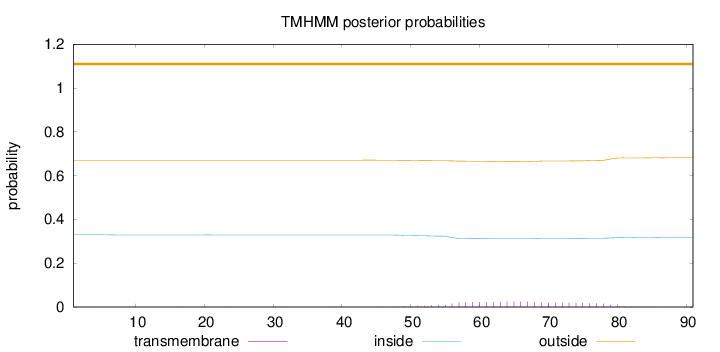
Topology
Subcellular location
Cell membrane
Length:
91
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.5745
Exp number, first 60 AAs:
0.16993
Total prob of N-in:
0.32960
outside
1 - 91
Population Genetic Test Statistics
Pi
46.327999
Theta
68.42386
Tajima's D
1.623887
CLR
2.946426
CSRT
0.817459127043648
Interpretation
Uncertain
