Gene
KWMTBOMO13267
Pre Gene Modal
BGIBMGA004733
Annotation
PREDICTED:_polypyrimidine_tract-binding_protein_2_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.136 Nuclear Reliability : 1.696
Sequence
CDS
ATGAAAACTAAGAGAGGCTCCGACGAGCTTCTTTCGCAGAGTGGCGTCACCGTCATGGCGCCCGCCGCTCCGCACCATGCCGACAATAACAATCAGGACTCTGCTGCCAAGAAGGTGAAACTGGAACAAAATGTCGGCAAGCCGTCTCGAGTCATTCACATACGGAACATACCGAACGAGACGACGGAAGCGGAGATCATACAACTCGGATTGCCCTTCGGAAGAGTCACCAACGTTCTAGTGCTCAAAGGCAAAAATCAGGCGTTCCTTGAAATGGCAGAGGAGATATCTGCGGTGGCTATGGTCGCATACTTCGGAGGATGTGTAGCGCAGTTGCGTGGCCGAGCGGTTTACGTTCAGTTCTCCAACCACAAGGAGCTAAAAACCGATCAGACGCACAGCAATGCCTCAGCATCGGCGCAAGCGGCTTTACAGGCGGCACAAGTCATATCGAATAGCGCTGGTGCGGCCATTGTTGCGGGCGGCGGTAACGCTTTGCAACCAGCTAATGGCGGTACAGAAATACAGGGCGGTCCCAATACAGTACTGCGTGTCATCATAGAGCATATGTTATATCCCATCGTTCTGGACGTCCTCTACTCGATCTTCCAACGCTACGGCAAGGTGCTCAAGATTGTAACTTTCACTAAAAACAATTCATTCCAAGCTCTCATTCAGTATCCAGACACGGTGTCGGCGCAAACAGCAAAAACGGCGCTCGACGGGCAGAATATCTACAACGGGTGCTGCACACTCCGCATCGACTATTCGAAGATGACTTGCCTGAACGTGAAATACAACAATGACAAGTCGCGGGACTACACGAATCCGACGCTTCCCTCCGGAGACGGCGACGCTCATCAGCTGTTGACCAGCGGAGGTGTATTGGCGCCTCAGTTCCTGGGCTTGGGTTCGGGATTGGCTTCGCCGTATGGTGTGGGTGTCGGTGGAGTGGGGCTCCCAGCCTTCGGAGCTCTCACTCTCACACCGGCCTCGCCCGCTCTGCGAGCCCTTGCTGCGCCACCTTTACCATTAGCCACAGTCCTACTGGTGTCTAACCTGAACGAAGAGATGGTCACGCCTGACGCTCTCTTTACCCTTTTCGGCGTGTATGGTGACGTGCAACGGGTGAAGATACTTTACAACAAGAAGGATTCGGCTTTGATTCAGATGGCAGAACCGCACCAGGCCCATCTAGCCCTGACCCACATGGACAAGTTACGCGTGTTCGGCAAGGCGATGCGCGTTATGCTAAGTAAGCACCAGACTGTTCAGCTCCCTAAAGAAGGACAACCTGACGCTGGTCTTACCAGGGACTACTCGCAGAGCCCACTCCACAGATTCAAGAAACCTGGTAGCAAAAATTACCAGAACATCTACCCGCCAAGCGCCACTCTGCATCTGTCGAACATTCCAGCCACCGTGACTGAAGAGGATATCAAAGATGCATTCTCCAAACGGGGATTCACAATTAAGGCTTTTAAATTTTTCCCGAAGGACCGCAAAATGGCTCTGGTCCAACTGTCGTCCATCGACGACGCAGTGACCGCGCTCATCAAGATGCACAACTACCAGCTCTCGGAATCCAATCACCTCAGGGTGTCTTTCTCCAAATCAAGCATCTAA
Protein
MKTKRGSDELLSQSGVTVMAPAAPHHADNNNQDSAAKKVKLEQNVGKPSRVIHIRNIPNETTEAEIIQLGLPFGRVTNVLVLKGKNQAFLEMAEEISAVAMVAYFGGCVAQLRGRAVYVQFSNHKELKTDQTHSNASASAQAALQAAQVISNSAGAAIVAGGGNALQPANGGTEIQGGPNTVLRVIIEHMLYPIVLDVLYSIFQRYGKVLKIVTFTKNNSFQALIQYPDTVSAQTAKTALDGQNIYNGCCTLRIDYSKMTCLNVKYNNDKSRDYTNPTLPSGDGDAHQLLTSGGVLAPQFLGLGSGLASPYGVGVGGVGLPAFGALTLTPASPALRALAAPPLPLATVLLVSNLNEEMVTPDALFTLFGVYGDVQRVKILYNKKDSALIQMAEPHQAHLALTHMDKLRVFGKAMRVMLSKHQTVQLPKEGQPDAGLTRDYSQSPLHRFKKPGSKNYQNIYPPSATLHLSNIPATVTEEDIKDAFSKRGFTIKAFKFFPKDRKMALVQLSSIDDAVTALIKMHNYQLSESNHLRVSFSKSSI
Summary
Uniprot
A0A3S2M470
A0A194R198
A0A2H1V283
A0A1Y1K2N2
A0A1Y1JZN1
A0A1Y1K016
+ More
A0A1Y1JZN6 A0A1Y1JZK8 A0A1Y1K7M1 A0A1Y1K7L2 A0A1Y1K2Q4 A0A1Y1K2P2 A0A1Y1K035 A0A1Y1JZL3 A0A1Y1K2P4 A0A212FB23 A0A139WBT4 K7J730 A0A1B6LM08 A0A1B6KR46 A0A1B6ETE8 A0A1B6HK58 A0A1B6HMP9 A0A1B6JVA8 A0A1B6HZH5 A0A1B6DRK8 A0A0C9QHP0 A0A1B6CQW8 A0A1B6CIA1 A0A0C9QWT6 A0A1B6LTS2 U5EZR3 A0A1B6CN94 A0A1Q3FTR8 A0A1Q3FTP4 W8B5A9 A0A1Q3FU40 W8AIY0 A0A1Q3FTK3 A0A1Q3FTY2 W8BFM8 A0A1Q3G1C1 A0A1Q3FTP1 A0A3R7PAJ9 W8AQZ2 A0A1Q3FTL4 W8AV00 A0A1Q3FU06 A0A1Q3G1C0 A0A034VSA2 A0A1J1I505 A0A0K8VCX3 A0A034VR94 A0A1I8M0Z5 A0A034VRA0 A0A0K8U7D6 A0A1W4VLU8 A0A1I8PG01 A0A194Q3K7 A0A0K8VZR3 A0A1W4VZY0 A0A1W4VM32 A0A1W4VYG7 B3LVE4 A0A0B4K6W9 B4IJ45 A0A0P9AN34 A0A0B4LHY1 B4QTB0 A0A0B4K6P4 Q8WR53 Q7KRS7 A0A0P8XWR8 A0A3B0KG51 Q95UI6 Q8IMF8 A8JRI1 A0A1I8PG87 T1P7L7 J9JUL5 A0A026WTC3 A0A0P6H4Q4 A0A1W4VZB7 A0A0A9ZC61 A0A0N8B8Q6 A0A0P5VQY5 Q16IC1 A0A1W4VYG3 A0A0Q9X083 A0A0Q9XCC7 A0A0A9WEP6 A0A0P5C910 A0A0K8SGN7 A0A0K8SA59 B4K859 A0A0P5C918 A0A0P8XWU6 E9GEU6 A0A0A9ZC66 Q7KES3
A0A1Y1JZN6 A0A1Y1JZK8 A0A1Y1K7M1 A0A1Y1K7L2 A0A1Y1K2Q4 A0A1Y1K2P2 A0A1Y1K035 A0A1Y1JZL3 A0A1Y1K2P4 A0A212FB23 A0A139WBT4 K7J730 A0A1B6LM08 A0A1B6KR46 A0A1B6ETE8 A0A1B6HK58 A0A1B6HMP9 A0A1B6JVA8 A0A1B6HZH5 A0A1B6DRK8 A0A0C9QHP0 A0A1B6CQW8 A0A1B6CIA1 A0A0C9QWT6 A0A1B6LTS2 U5EZR3 A0A1B6CN94 A0A1Q3FTR8 A0A1Q3FTP4 W8B5A9 A0A1Q3FU40 W8AIY0 A0A1Q3FTK3 A0A1Q3FTY2 W8BFM8 A0A1Q3G1C1 A0A1Q3FTP1 A0A3R7PAJ9 W8AQZ2 A0A1Q3FTL4 W8AV00 A0A1Q3FU06 A0A1Q3G1C0 A0A034VSA2 A0A1J1I505 A0A0K8VCX3 A0A034VR94 A0A1I8M0Z5 A0A034VRA0 A0A0K8U7D6 A0A1W4VLU8 A0A1I8PG01 A0A194Q3K7 A0A0K8VZR3 A0A1W4VZY0 A0A1W4VM32 A0A1W4VYG7 B3LVE4 A0A0B4K6W9 B4IJ45 A0A0P9AN34 A0A0B4LHY1 B4QTB0 A0A0B4K6P4 Q8WR53 Q7KRS7 A0A0P8XWR8 A0A3B0KG51 Q95UI6 Q8IMF8 A8JRI1 A0A1I8PG87 T1P7L7 J9JUL5 A0A026WTC3 A0A0P6H4Q4 A0A1W4VZB7 A0A0A9ZC61 A0A0N8B8Q6 A0A0P5VQY5 Q16IC1 A0A1W4VYG3 A0A0Q9X083 A0A0Q9XCC7 A0A0A9WEP6 A0A0P5C910 A0A0K8SGN7 A0A0K8SA59 B4K859 A0A0P5C918 A0A0P8XWU6 E9GEU6 A0A0A9ZC66 Q7KES3
Pubmed
EMBL
RSAL01000040
RVE50963.1
KQ460883
KPJ11289.1
ODYU01000346
SOQ34941.1
+ More
GEZM01096618 JAV54758.1 GEZM01096608 JAV54772.1 GEZM01096622 JAV54753.1 GEZM01096606 JAV54774.1 GEZM01096621 JAV54754.1 GEZM01096612 JAV54767.1 GEZM01096619 JAV54757.1 GEZM01096609 JAV54771.1 GEZM01096611 JAV54768.1 GEZM01096607 JAV54773.1 GEZM01096614 JAV54764.1 GEZM01096616 JAV54761.1 AGBW02009387 OWR50936.1 KQ971372 KYB25386.1 AAZX01002966 AAZX01005944 AAZX01008283 AAZX01008561 AAZX01010264 AAZX01013204 AAZX01017040 AAZX01017275 AAZX01020952 AAZX01022065 GEBQ01015289 JAT24688.1 GEBQ01026069 JAT13908.1 GECZ01028540 JAS41229.1 GECU01032649 JAS75057.1 GECU01031738 JAS75968.1 GECU01004555 JAT03152.1 GECU01027637 JAS80069.1 GEDC01008993 JAS28305.1 GBYB01003049 JAG72816.1 GEDC01021613 JAS15685.1 GEDC01024243 JAS13055.1 GBYB01008194 JAG77961.1 GEBQ01012920 JAT27057.1 GANO01000309 JAB59562.1 GEDC01022440 JAS14858.1 GFDL01004107 JAV30938.1 GFDL01004110 JAV30935.1 GAMC01018089 JAB88466.1 GFDL01004102 JAV30943.1 GAMC01018090 JAB88465.1 GFDL01004104 JAV30941.1 GFDL01004109 JAV30936.1 GAMC01018086 JAB88469.1 GFDL01001448 JAV33597.1 GFDL01004108 JAV30937.1 QCYY01003104 ROT65304.1 GAMC01018088 JAB88467.1 GFDL01004105 JAV30940.1 GAMC01018087 JAB88468.1 GFDL01004103 JAV30942.1 GFDL01001452 JAV33593.1 GAKP01013955 JAC44997.1 CVRI01000040 CRK94842.1 GDHF01015919 GDHF01009131 GDHF01008096 GDHF01007263 JAI36395.1 JAI43183.1 JAI44218.1 JAI45051.1 GAKP01013959 GAKP01013958 GAKP01013957 GAKP01013956 GAKP01013954 JAC44994.1 GAKP01013953 JAC44999.1 GDHF01030089 GDHF01019940 JAI22225.1 JAI32374.1 KQ459586 KPI97995.1 GDHF01007988 GDHF01004259 JAI44326.1 JAI48055.1 CH902617 EDV41472.2 AE014297 AFH06716.1 CH480846 EDW50998.1 KPU79170.1 AHN57629.1 CM000364 EDX15172.1 AFH06715.1 AF455053 BT032832 AAF57208.2 AAL57860.1 AAO41621.1 AAO41622.1 AAO41624.1 AAO41625.1 AAO41626.1 ABW08818.1 ACD81846.1 ACZ95081.1 ACZ95087.1 AAO41623.2 KPU79171.1 OUUW01000007 SPP82618.1 AY052367 BT030984 AAL14775.1 AAO41627.1 ABV82366.1 ACZ95080.1 ACZ95083.1 AF211191 AAF22979.1 AAN14296.1 ACZ95084.1 ACZ95085.1 ACZ95086.1 ABW08816.1 KA644464 AFP59093.1 ABLF02032856 ABLF02032857 KK107109 EZA59198.1 GDIQ01255288 GDIQ01034664 JAN60073.1 GBHO01000752 JAG42852.1 GDIQ01201811 JAK49914.1 GDIQ01201812 GDIP01096752 JAK49913.1 JAM06963.1 CH478088 EAT34016.1 CH933806 KRG01542.1 KRG01544.1 GBHO01036682 GBHO01036679 GDHC01021097 JAG06922.1 JAG06925.1 JAP97531.1 GDIP01173814 JAJ49588.1 GBRD01013373 GBRD01013369 JAG52457.1 GBRD01016186 GBRD01008475 JAG49640.1 EDW15413.2 GDIP01173813 JAJ49589.1 KPU79169.1 GL732541 EFX81898.1 GBHO01000747 JAG42857.1 AF436844 AAL27010.1 ABW08817.1
GEZM01096618 JAV54758.1 GEZM01096608 JAV54772.1 GEZM01096622 JAV54753.1 GEZM01096606 JAV54774.1 GEZM01096621 JAV54754.1 GEZM01096612 JAV54767.1 GEZM01096619 JAV54757.1 GEZM01096609 JAV54771.1 GEZM01096611 JAV54768.1 GEZM01096607 JAV54773.1 GEZM01096614 JAV54764.1 GEZM01096616 JAV54761.1 AGBW02009387 OWR50936.1 KQ971372 KYB25386.1 AAZX01002966 AAZX01005944 AAZX01008283 AAZX01008561 AAZX01010264 AAZX01013204 AAZX01017040 AAZX01017275 AAZX01020952 AAZX01022065 GEBQ01015289 JAT24688.1 GEBQ01026069 JAT13908.1 GECZ01028540 JAS41229.1 GECU01032649 JAS75057.1 GECU01031738 JAS75968.1 GECU01004555 JAT03152.1 GECU01027637 JAS80069.1 GEDC01008993 JAS28305.1 GBYB01003049 JAG72816.1 GEDC01021613 JAS15685.1 GEDC01024243 JAS13055.1 GBYB01008194 JAG77961.1 GEBQ01012920 JAT27057.1 GANO01000309 JAB59562.1 GEDC01022440 JAS14858.1 GFDL01004107 JAV30938.1 GFDL01004110 JAV30935.1 GAMC01018089 JAB88466.1 GFDL01004102 JAV30943.1 GAMC01018090 JAB88465.1 GFDL01004104 JAV30941.1 GFDL01004109 JAV30936.1 GAMC01018086 JAB88469.1 GFDL01001448 JAV33597.1 GFDL01004108 JAV30937.1 QCYY01003104 ROT65304.1 GAMC01018088 JAB88467.1 GFDL01004105 JAV30940.1 GAMC01018087 JAB88468.1 GFDL01004103 JAV30942.1 GFDL01001452 JAV33593.1 GAKP01013955 JAC44997.1 CVRI01000040 CRK94842.1 GDHF01015919 GDHF01009131 GDHF01008096 GDHF01007263 JAI36395.1 JAI43183.1 JAI44218.1 JAI45051.1 GAKP01013959 GAKP01013958 GAKP01013957 GAKP01013956 GAKP01013954 JAC44994.1 GAKP01013953 JAC44999.1 GDHF01030089 GDHF01019940 JAI22225.1 JAI32374.1 KQ459586 KPI97995.1 GDHF01007988 GDHF01004259 JAI44326.1 JAI48055.1 CH902617 EDV41472.2 AE014297 AFH06716.1 CH480846 EDW50998.1 KPU79170.1 AHN57629.1 CM000364 EDX15172.1 AFH06715.1 AF455053 BT032832 AAF57208.2 AAL57860.1 AAO41621.1 AAO41622.1 AAO41624.1 AAO41625.1 AAO41626.1 ABW08818.1 ACD81846.1 ACZ95081.1 ACZ95087.1 AAO41623.2 KPU79171.1 OUUW01000007 SPP82618.1 AY052367 BT030984 AAL14775.1 AAO41627.1 ABV82366.1 ACZ95080.1 ACZ95083.1 AF211191 AAF22979.1 AAN14296.1 ACZ95084.1 ACZ95085.1 ACZ95086.1 ABW08816.1 KA644464 AFP59093.1 ABLF02032856 ABLF02032857 KK107109 EZA59198.1 GDIQ01255288 GDIQ01034664 JAN60073.1 GBHO01000752 JAG42852.1 GDIQ01201811 JAK49914.1 GDIQ01201812 GDIP01096752 JAK49913.1 JAM06963.1 CH478088 EAT34016.1 CH933806 KRG01542.1 KRG01544.1 GBHO01036682 GBHO01036679 GDHC01021097 JAG06922.1 JAG06925.1 JAP97531.1 GDIP01173814 JAJ49588.1 GBRD01013373 GBRD01013369 JAG52457.1 GBRD01016186 GBRD01008475 JAG49640.1 EDW15413.2 GDIP01173813 JAJ49589.1 KPU79169.1 GL732541 EFX81898.1 GBHO01000747 JAG42857.1 AF436844 AAL27010.1 ABW08817.1
Proteomes
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
A0A3S2M470
A0A194R198
A0A2H1V283
A0A1Y1K2N2
A0A1Y1JZN1
A0A1Y1K016
+ More
A0A1Y1JZN6 A0A1Y1JZK8 A0A1Y1K7M1 A0A1Y1K7L2 A0A1Y1K2Q4 A0A1Y1K2P2 A0A1Y1K035 A0A1Y1JZL3 A0A1Y1K2P4 A0A212FB23 A0A139WBT4 K7J730 A0A1B6LM08 A0A1B6KR46 A0A1B6ETE8 A0A1B6HK58 A0A1B6HMP9 A0A1B6JVA8 A0A1B6HZH5 A0A1B6DRK8 A0A0C9QHP0 A0A1B6CQW8 A0A1B6CIA1 A0A0C9QWT6 A0A1B6LTS2 U5EZR3 A0A1B6CN94 A0A1Q3FTR8 A0A1Q3FTP4 W8B5A9 A0A1Q3FU40 W8AIY0 A0A1Q3FTK3 A0A1Q3FTY2 W8BFM8 A0A1Q3G1C1 A0A1Q3FTP1 A0A3R7PAJ9 W8AQZ2 A0A1Q3FTL4 W8AV00 A0A1Q3FU06 A0A1Q3G1C0 A0A034VSA2 A0A1J1I505 A0A0K8VCX3 A0A034VR94 A0A1I8M0Z5 A0A034VRA0 A0A0K8U7D6 A0A1W4VLU8 A0A1I8PG01 A0A194Q3K7 A0A0K8VZR3 A0A1W4VZY0 A0A1W4VM32 A0A1W4VYG7 B3LVE4 A0A0B4K6W9 B4IJ45 A0A0P9AN34 A0A0B4LHY1 B4QTB0 A0A0B4K6P4 Q8WR53 Q7KRS7 A0A0P8XWR8 A0A3B0KG51 Q95UI6 Q8IMF8 A8JRI1 A0A1I8PG87 T1P7L7 J9JUL5 A0A026WTC3 A0A0P6H4Q4 A0A1W4VZB7 A0A0A9ZC61 A0A0N8B8Q6 A0A0P5VQY5 Q16IC1 A0A1W4VYG3 A0A0Q9X083 A0A0Q9XCC7 A0A0A9WEP6 A0A0P5C910 A0A0K8SGN7 A0A0K8SA59 B4K859 A0A0P5C918 A0A0P8XWU6 E9GEU6 A0A0A9ZC66 Q7KES3
A0A1Y1JZN6 A0A1Y1JZK8 A0A1Y1K7M1 A0A1Y1K7L2 A0A1Y1K2Q4 A0A1Y1K2P2 A0A1Y1K035 A0A1Y1JZL3 A0A1Y1K2P4 A0A212FB23 A0A139WBT4 K7J730 A0A1B6LM08 A0A1B6KR46 A0A1B6ETE8 A0A1B6HK58 A0A1B6HMP9 A0A1B6JVA8 A0A1B6HZH5 A0A1B6DRK8 A0A0C9QHP0 A0A1B6CQW8 A0A1B6CIA1 A0A0C9QWT6 A0A1B6LTS2 U5EZR3 A0A1B6CN94 A0A1Q3FTR8 A0A1Q3FTP4 W8B5A9 A0A1Q3FU40 W8AIY0 A0A1Q3FTK3 A0A1Q3FTY2 W8BFM8 A0A1Q3G1C1 A0A1Q3FTP1 A0A3R7PAJ9 W8AQZ2 A0A1Q3FTL4 W8AV00 A0A1Q3FU06 A0A1Q3G1C0 A0A034VSA2 A0A1J1I505 A0A0K8VCX3 A0A034VR94 A0A1I8M0Z5 A0A034VRA0 A0A0K8U7D6 A0A1W4VLU8 A0A1I8PG01 A0A194Q3K7 A0A0K8VZR3 A0A1W4VZY0 A0A1W4VM32 A0A1W4VYG7 B3LVE4 A0A0B4K6W9 B4IJ45 A0A0P9AN34 A0A0B4LHY1 B4QTB0 A0A0B4K6P4 Q8WR53 Q7KRS7 A0A0P8XWR8 A0A3B0KG51 Q95UI6 Q8IMF8 A8JRI1 A0A1I8PG87 T1P7L7 J9JUL5 A0A026WTC3 A0A0P6H4Q4 A0A1W4VZB7 A0A0A9ZC61 A0A0N8B8Q6 A0A0P5VQY5 Q16IC1 A0A1W4VYG3 A0A0Q9X083 A0A0Q9XCC7 A0A0A9WEP6 A0A0P5C910 A0A0K8SGN7 A0A0K8SA59 B4K859 A0A0P5C918 A0A0P8XWU6 E9GEU6 A0A0A9ZC66 Q7KES3
PDB
2MJU
E-value=6.91991e-73,
Score=698
Ontologies
GO
Topology
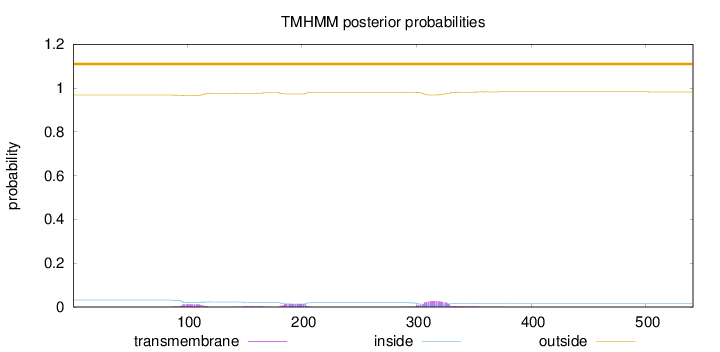
Length:
541
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.32776
Exp number, first 60 AAs:
0.0003
Total prob of N-in:
0.03193
outside
1 - 541
Population Genetic Test Statistics
Pi
229.039213
Theta
175.946839
Tajima's D
0.458216
CLR
0.567002
CSRT
0.499725013749313
Interpretation
Uncertain
