Gene
KWMTBOMO13249 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004726
Annotation
PREDICTED:_antichymotrypsin-2-like_[Bombyx_mori]
Full name
Antichymotrypsin-2
Alternative Name
Antichymotrypsin II
Location in the cell
Extracellular Reliability : 2.41
Sequence
CDS
ATGCCTTATAACTCATGCAATGGGGATCCGAACGCTGTTGCTGGATGTGGAGTAAATTGTAACAGACATTGTTCTGATATCGGCAAAGAATGTCAAAGTTGTGCTGCTGTTTGCTACGAAGACGGCTGCGATTGTAAAGACGGATATTATTACGATGATAACATTAATAAATGTGTTTTACCCGAAGACTGCACTCCTACTTGTGGTAATGATGAAGTCTACAATGATTGTATACAAGGGTACTGCCAACCGAAGAATTGTTCGGAAATAGGGAAACCCGTTGCTTGCCCAAGAATAGATCCCAAGAATTGCATCAAAGGGTGCTTGTGTAAAGAAGACTACGTCAGAGCTGATAATGGAACGTGTATACCAAAAATCGACTGTCCTACTACCAATTCATCGGGAAATGTTGACCAATCCTTGGAAAAACTCCGCAGAGGAAACATGGCATTAGTTGGCCAGTTTCTTTGGGAATTATACAAAATGAATCCAGGACAAAGTTTCGTTACATCACCCGTATCTGTACTCATTCCTTACGGTCAACTAGCGCTATACGCCGTTAGAGAAACTTTAGATCAACTATTAAAGTTTATAAACCTCGACAATAAGGATCAGATCAAGGAAGCTTTCCCCGCTTTGCTTACGAATTATCGGTCTCAAAGCTCAGTTCTTTTGACGGTGGCAGTTAAATGCTACGGCAACATAAATTATCCTTTTACGGACCAGTTTAAGAACGACGCTGTAACTTACTTTGACTCAGAAGGAGAAAATATTGATTTTAAATATGCGAAAGAAGCTGCAGACACAATAAACGGATGGGTCGCAAACAAAACTAATAACTTAATTCAAAATCTTGTCAGCGAAGATTTGTTCAGCGATGACACTAGATTGGTTCTAGTTAACGGCATTTATTTCCTGGGAAATTGGCAAAATCAGTTCAATCCTAATAACACTCGCGATAGAGACTTCTACATCACACCAAACAAAACTAAGTCCATAAAGACTATGTATCAAGAAAACAACTTTAATTATGGAGAAAACGAGATTCTCGATGCACAGATCTTAGAATTGTTCTACAAAGGCGGCAATTCTAGTTTAGTAATCTTGTTACCAAAGAAAAAAGATGGCCTTCTCCAGATGTTAGATAAATTACGAAATTCAGATGAATTATCTAATAGCATTAAATCACTTAGGCAAGAGAAAGTCAAATGTTTCCTACCTAAGATGGATATAGAAACGAACATCGATATAGCAGAACTTTCCCAGAAGCTGAATGTAACCAAAATGTTCGATCCACGATCAGATGATTTCGAAGGAATTTTATTAAATTCGGACCCTGTATATGTTTCTGCTGCAATACAGAAAGCTAAAATCATAGTTGATGAAACTGGAACCGAGGCAGCGGCAGCAACTGTTATACTTCTGTACTCATTTAGAAATGCATTTGATCCTTTATTTGTTCAACAAGATATTTTGGATTCAGATTTACAAGACCATCCACACGTATTCTTCGTTTTGATTAACGATGAACCAATTTTCAGTGGATTGTTCATAGGTGCCTAA
Protein
MPYNSCNGDPNAVAGCGVNCNRHCSDIGKECQSCAAVCYEDGCDCKDGYYYDDNINKCVLPEDCTPTCGNDEVYNDCIQGYCQPKNCSEIGKPVACPRIDPKNCIKGCLCKEDYVRADNGTCIPKIDCPTTNSSGNVDQSLEKLRRGNMALVGQFLWELYKMNPGQSFVTSPVSVLIPYGQLALYAVRETLDQLLKFINLDNKDQIKEAFPALLTNYRSQSSVLLTVAVKCYGNINYPFTDQFKNDAVTYFDSEGENIDFKYAKEAADTINGWVANKTNNLIQNLVSEDLFSDDTRLVLVNGIYFLGNWQNQFNPNNTRDRDFYITPNKTKSIKTMYQENNFNYGENEILDAQILELFYKGGNSSLVILLPKKKDGLLQMLDKLRNSDELSNSIKSLRQEKVKCFLPKMDIETNIDIAELSQKLNVTKMFDPRSDDFEGILLNSDPVYVSAAIQKAKIIVDETGTEAAAATVILLYSFRNAFDPLFVQQDILDSDLQDHPHVFFVLINDEPIFSGLFIGA
Summary
Miscellaneous
The N-terminal section (1-336) is identical to the N-terminal section of the silk moth antitrypsin I (17-352).
Similarity
Belongs to the serpin family.
Keywords
Complete proteome
Direct protein sequencing
Protease inhibitor
Reference proteome
Secreted
Serine protease inhibitor
Feature
chain Antichymotrypsin-2
Uniprot
A0A3G1T1Q1
A0A2A4J7B9
G9F9J2
H9J5D8
A0A3S2M6W8
G9F9J3
+ More
A0A3G1T1R3 A0A194R0R3 A0A1E1WCC9 A0A194PXE0 A0A1E1WM18 A0A1E1W9X2 A0A212EGT4 A0A3S2NJW5 K4P392 Q25492 A3KEZ7 Q25501 Q60FS0 Q25495 Q25500 B4XH99 F5B4G8 Q25498 I4DJC8 J7HI16 A0A194PZ13 C7ASM4 A0A2H1VE36 C7ASM3 C7ASM6 A0A3G1T1E7 P80034 C7ASM7 B0FZ61 C5I794 A0A0M4K7J2 Q8IS86 B0FZ64 B0FZ60 B0FZ62 B0FZ63 A0A212FD15 A0A2A4JQC6 A0A2A4JPB5 Q8MPN5 Q8MPN7
A0A3G1T1R3 A0A194R0R3 A0A1E1WCC9 A0A194PXE0 A0A1E1WM18 A0A1E1W9X2 A0A212EGT4 A0A3S2NJW5 K4P392 Q25492 A3KEZ7 Q25501 Q60FS0 Q25495 Q25500 B4XH99 F5B4G8 Q25498 I4DJC8 J7HI16 A0A194PZ13 C7ASM4 A0A2H1VE36 C7ASM3 C7ASM6 A0A3G1T1E7 P80034 C7ASM7 B0FZ61 C5I794 A0A0M4K7J2 Q8IS86 B0FZ64 B0FZ60 B0FZ62 B0FZ63 A0A212FD15 A0A2A4JQC6 A0A2A4JPB5 Q8MPN5 Q8MPN7
Pubmed
EMBL
MG770320
AXY94922.1
NWSH01002542
PCG68017.1
JN033737
AEW46890.2
+ More
BABH01039203 BABH01039204 BABH01039205 BABH01039206 BABH01039207 RSAL01000018 RVE52848.1 JN033738 AEW46891.2 MG770321 AXY94923.1 KQ460883 KPJ11292.1 GDQN01006457 JAT84597.1 KQ459586 KPI97992.1 GDQN01003022 JAT88032.1 GDQN01007288 JAT83766.1 AGBW02015025 OWR40695.1 RVE52846.1 JX524148 AFV46312.1 U58361 AAC47338.1 AB295498 BAF48334.1 AAC47339.1 AB189029 BAD52261.1 AAC47341.1 AAC47335.1 EU040208 ABW17156.1 HQ615869 ADT63775.1 AAC47342.1 AK401396 BAM18018.1 JN400442 AFQ01141.1 KPI97989.1 FJ613795 ACT36274.1 ODYU01002062 SOQ39103.1 FJ613794 FJ613796 ACT36273.1 ACT36275.1 FJ613797 ACT36278.1 MG992370 AXY94808.1 ACT36277.1 ACT36279.1 EU368832 ABY68562.1 FJ763763 ACR56864.1 KR003729 ALD62507.1 AY148483 AAN71632.1 ABY68559.1 ABY68563.1 ABY68561.1 ABY68560.1 AGBW02009121 OWR51607.1 NWSH01000840 PCG73908.1 PCG73905.1 AE013599 AJ428889 AAS64783.1 CAD21901.1 BT125838 AJ428884 AAS64782.1 ADR83723.1 CAD21896.1
BABH01039203 BABH01039204 BABH01039205 BABH01039206 BABH01039207 RSAL01000018 RVE52848.1 JN033738 AEW46891.2 MG770321 AXY94923.1 KQ460883 KPJ11292.1 GDQN01006457 JAT84597.1 KQ459586 KPI97992.1 GDQN01003022 JAT88032.1 GDQN01007288 JAT83766.1 AGBW02015025 OWR40695.1 RVE52846.1 JX524148 AFV46312.1 U58361 AAC47338.1 AB295498 BAF48334.1 AAC47339.1 AB189029 BAD52261.1 AAC47341.1 AAC47335.1 EU040208 ABW17156.1 HQ615869 ADT63775.1 AAC47342.1 AK401396 BAM18018.1 JN400442 AFQ01141.1 KPI97989.1 FJ613795 ACT36274.1 ODYU01002062 SOQ39103.1 FJ613794 FJ613796 ACT36273.1 ACT36275.1 FJ613797 ACT36278.1 MG992370 AXY94808.1 ACT36277.1 ACT36279.1 EU368832 ABY68562.1 FJ763763 ACR56864.1 KR003729 ALD62507.1 AY148483 AAN71632.1 ABY68559.1 ABY68563.1 ABY68561.1 ABY68560.1 AGBW02009121 OWR51607.1 NWSH01000840 PCG73908.1 PCG73905.1 AE013599 AJ428889 AAS64783.1 CAD21901.1 BT125838 AJ428884 AAS64782.1 ADR83723.1 CAD21896.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A3G1T1Q1
A0A2A4J7B9
G9F9J2
H9J5D8
A0A3S2M6W8
G9F9J3
+ More
A0A3G1T1R3 A0A194R0R3 A0A1E1WCC9 A0A194PXE0 A0A1E1WM18 A0A1E1W9X2 A0A212EGT4 A0A3S2NJW5 K4P392 Q25492 A3KEZ7 Q25501 Q60FS0 Q25495 Q25500 B4XH99 F5B4G8 Q25498 I4DJC8 J7HI16 A0A194PZ13 C7ASM4 A0A2H1VE36 C7ASM3 C7ASM6 A0A3G1T1E7 P80034 C7ASM7 B0FZ61 C5I794 A0A0M4K7J2 Q8IS86 B0FZ64 B0FZ60 B0FZ62 B0FZ63 A0A212FD15 A0A2A4JQC6 A0A2A4JPB5 Q8MPN5 Q8MPN7
A0A3G1T1R3 A0A194R0R3 A0A1E1WCC9 A0A194PXE0 A0A1E1WM18 A0A1E1W9X2 A0A212EGT4 A0A3S2NJW5 K4P392 Q25492 A3KEZ7 Q25501 Q60FS0 Q25495 Q25500 B4XH99 F5B4G8 Q25498 I4DJC8 J7HI16 A0A194PZ13 C7ASM4 A0A2H1VE36 C7ASM3 C7ASM6 A0A3G1T1E7 P80034 C7ASM7 B0FZ61 C5I794 A0A0M4K7J2 Q8IS86 B0FZ64 B0FZ60 B0FZ62 B0FZ63 A0A212FD15 A0A2A4JQC6 A0A2A4JPB5 Q8MPN5 Q8MPN7
PDB
1SEK
E-value=5.68576e-59,
Score=578
Ontologies
PANTHER
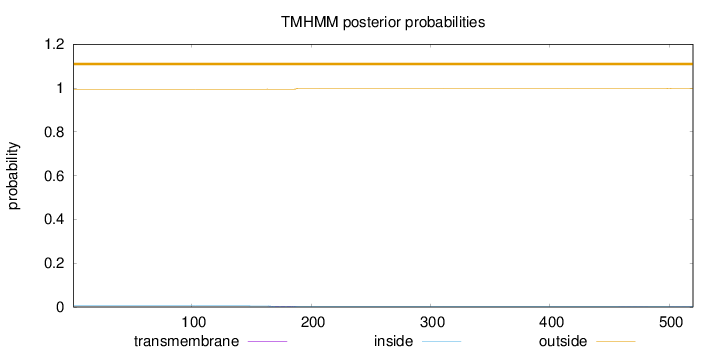
Topology
Subcellular location
Secreted
Length:
520
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10772
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00514
outside
1 - 520
Population Genetic Test Statistics
Pi
153.293792
Theta
19.733854
Tajima's D
0
CLR
1.561678
CSRT
0.368831558422079
Interpretation
Uncertain
