Gene
KWMTBOMO13234
Pre Gene Modal
BGIBMGA009973
Annotation
PREDICTED:_carbohydrate_sulfotransferase_11_[Papilio_xuthus]
Full name
Carbohydrate sulfotransferase
Location in the cell
Nuclear Reliability : 1.91
Sequence
CDS
ATGACAATTTTCAAGAGCAACACCACAACTGCACCGGTGAAGGATTGGGCGATACCACCTGATAATTACACCAAATGGATGTTTCAGGGTCCAGTTATAGGCGATGACGAGCCTGAGACGGATTCGAATCAGAACCAATGGTTGGAGCCAGACAACATAACGGTCCAAGAACTAGAGCAACGCGTAATTAGAGTACAGAATGTTTGTCGTAAGCGACACCTGCCTTCAGATACAATAAATAACAAGGAATTCTTTGTAGATCACCTGCATAATCTCGTGTGGTGTAATGTTTTTAAAGCTGCCAGTTCCTCTTGGTTATACAAATTCAACATTCTAGGCGGTTACGATAGGAAATTTTTGGGAAGAACAAGGCAAACGCCTCTGATGCTGGCAAGAAAAAAGTTTCCACGTCCAAATGCCGAAGAATTGAAGGACTCTATAAACACGCCCGGCGTAATTTCTTTAATGATAGTTCGCGAGCCATTCGTTCGTCTCCTGTCAGCGTATCGGGATAAATTAGAGAACATAACTCCGCCATATTACAGAAAACTAGCGAGGGCCATCGTGGCCAGGCACAGGGAAGATGCAACAAAAATCCTTGGACCAATAAAAAGTTTCGGACCTACGTTTTACGAATTCGTAGCTTATTTGATACATAAACATAAGGAAAAAGATGAAAAATTTACTTTTGATGAGCACTGGGCGCCGTATTATCAGTTTTGCTCTCCGTGCGCAGTTAATTTTACAGTAATCGGTAAAGTGGAAACTCTTAAAAGGGACTCTGCGTATGTGATACAACAACTAGGTCTCGGACATTTGTTAGGTCGAACCACAGGGGATAAGCGGACGAGACTTCGTACAGTAATGAACAAATCAAGAGACGGAAAGAACACGACATCTATATTGAAATATTACTTTAGTCAACTAAATGAAAATTTGCTTAAAGATCTCTTGAATATTTACGGTATAGATTTCGAAATGTTTGGATATGATCCAAATGTGTATCAAAGTTTCGTTAGAAAATAA
Protein
MTIFKSNTTTAPVKDWAIPPDNYTKWMFQGPVIGDDEPETDSNQNQWLEPDNITVQELEQRVIRVQNVCRKRHLPSDTINNKEFFVDHLHNLVWCNVFKAASSSWLYKFNILGGYDRKFLGRTRQTPLMLARKKFPRPNAEELKDSINTPGVISLMIVREPFVRLLSAYRDKLENITPPYYRKLARAIVARHREDATKILGPIKSFGPTFYEFVAYLIHKHKEKDEKFTFDEHWAPYYQFCSPCAVNFTVIGKVETLKRDSAYVIQQLGLGHLLGRTTGDKRTRLRTVMNKSRDGKNTTSILKYYFSQLNENLLKDLLNIYGIDFEMFGYDPNVYQSFVRK
Summary
Similarity
Belongs to the sulfotransferase 2 family.
Uniprot
H9JKC2
A0A194PXB2
A0A194R0I1
A0A212ER84
A0A0L7LPL0
A0A2H1VI30
+ More
A0A2J7Q7Y6 A0A2P8XIU4 A0A067RLT5 A0A0L0C9C5 Q16TU6 A0A182GUH3 B3MLL8 A0A1W4UWA5 A0A182YFD2 B4Q3K6 B3N4U7 B4I1F1 A0A182SFU7 Q9VMS3 B4P075 A0A3B0JB11 A0A182MUA9 A0A182QLS7 A0A1J1HZT2 B4KI39 Q7PW21 A0A182V690 A0A182R9M9 A0A0R3NTN8 A0A182WH09 A0A1B0BES2 Q29M12 A0A182JML7 A0A1A9XRX3 B4G6Q7 A0A182KUN5 A0A182NEY8 A0A182U842 A0A1A9WUW0 A0A182WYR3 B4N0Z4 B4LRA7 A0A182I508 A0A1S4H0Z4 A0A1B0D8W1 A0A1B0FFN9 B0W1J7 W5JS76 A0A1A9ZK91 A0A0M4ENM4 A0A182F5F3 A0A1B6DBS3 A0A1A9VXY1 B4JPK6 A0A1B0CW14 A0A1I8PV94 A0A1I8MI65 A0A336LSE2 A0A067QKN1 A0A1B0CY71 A0A182S9P5 A0A182MSR1 A0A1W4WQX9 A0A182WKU9 A0A182YD58 A0A0K8TKD1 A0A182RIJ1 N6T5B1 A0A1Y1KP93 A0A182NIC2 D6WTL6 A0A084WDM9 A0A0C9QSZ1 A0A146M8T9 A0A0A9X699 A0A1B6CEK6 A0A182XMR2 A0A2A3EK66 A0A182IS71 V9IIL2 A0A182UV54 A0A182FE55 A0A023ETD2 A0A1J1J607 B0W401 A0A182QLH1 A0A182L309 A0A182HWM0 Q7Q6Q9 A0A3R7M829 A0A195BQC1 F4W9V1 A0A151WL04 A0A1B6EP42 A0A1B6J036 A0A1B6GMP7
A0A2J7Q7Y6 A0A2P8XIU4 A0A067RLT5 A0A0L0C9C5 Q16TU6 A0A182GUH3 B3MLL8 A0A1W4UWA5 A0A182YFD2 B4Q3K6 B3N4U7 B4I1F1 A0A182SFU7 Q9VMS3 B4P075 A0A3B0JB11 A0A182MUA9 A0A182QLS7 A0A1J1HZT2 B4KI39 Q7PW21 A0A182V690 A0A182R9M9 A0A0R3NTN8 A0A182WH09 A0A1B0BES2 Q29M12 A0A182JML7 A0A1A9XRX3 B4G6Q7 A0A182KUN5 A0A182NEY8 A0A182U842 A0A1A9WUW0 A0A182WYR3 B4N0Z4 B4LRA7 A0A182I508 A0A1S4H0Z4 A0A1B0D8W1 A0A1B0FFN9 B0W1J7 W5JS76 A0A1A9ZK91 A0A0M4ENM4 A0A182F5F3 A0A1B6DBS3 A0A1A9VXY1 B4JPK6 A0A1B0CW14 A0A1I8PV94 A0A1I8MI65 A0A336LSE2 A0A067QKN1 A0A1B0CY71 A0A182S9P5 A0A182MSR1 A0A1W4WQX9 A0A182WKU9 A0A182YD58 A0A0K8TKD1 A0A182RIJ1 N6T5B1 A0A1Y1KP93 A0A182NIC2 D6WTL6 A0A084WDM9 A0A0C9QSZ1 A0A146M8T9 A0A0A9X699 A0A1B6CEK6 A0A182XMR2 A0A2A3EK66 A0A182IS71 V9IIL2 A0A182UV54 A0A182FE55 A0A023ETD2 A0A1J1J607 B0W401 A0A182QLH1 A0A182L309 A0A182HWM0 Q7Q6Q9 A0A3R7M829 A0A195BQC1 F4W9V1 A0A151WL04 A0A1B6EP42 A0A1B6J036 A0A1B6GMP7
EC Number
2.8.2.-
Pubmed
19121390
26354079
22118469
26227816
29403074
24845553
+ More
26108605 17510324 26483478 17994087 25244985 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 12364791 15632085 20966253 20920257 23761445 25315136 26369729 23537049 28004739 18362917 19820115 24438588 26823975 25401762 24945155 14747013 17210077 21719571
26108605 17510324 26483478 17994087 25244985 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 12364791 15632085 20966253 20920257 23761445 25315136 26369729 23537049 28004739 18362917 19820115 24438588 26823975 25401762 24945155 14747013 17210077 21719571
EMBL
BABH01036744
KQ459586
KPI97986.1
KQ460883
KPJ11298.1
AGBW02013113
+ More
OWR43961.1 JTDY01000397 KOB77370.1 ODYU01002682 SOQ40495.1 NEVH01017000 PNF24697.1 PYGN01001983 PSN31927.1 KK852601 KDR20510.1 JRES01000714 KNC29023.1 CH477639 EAT37943.1 JXUM01089185 JXUM01089186 KQ563751 KXJ73378.1 CH902620 EDV31767.1 CM000361 CM002910 EDX03810.1 KMY88279.1 CH954177 EDV57849.1 CH480820 EDW54358.1 AE014134 AY051453 AAF52238.1 AAK92877.1 AGB92630.1 CM000157 EDW88940.1 OUUW01000004 SPP79537.1 AXCM01001573 AXCN02000839 CVRI01000037 CRK93608.1 CH933807 EDW12332.1 AAAB01008984 EAA15163.4 CH379060 KRT04300.1 JXJN01012999 EAL33883.3 CH479180 EDW29171.1 CH963920 EDW78156.1 CH940649 EDW64577.1 APCN01003745 AJVK01004152 AJVK01004153 AJVK01004154 CCAG010014402 CCAG010014403 CCAG010014404 DS231822 EDS25795.1 ADMH02000581 ETN65775.1 CP012523 ALC38287.1 GEDC01014208 JAS23090.1 CH916372 EDV98836.1 AJWK01031561 UFQT01000144 SSX20840.1 KK853243 KDR09457.1 AJWK01035509 AXCM01006480 AXCM01006481 GDAI01002816 JAI14787.1 APGK01014012 APGK01052024 KB741208 KB739171 KB631654 ENN72858.1 ENN82357.1 ERL85063.1 GEZM01077621 GEZM01077620 JAV63233.1 KQ971355 EFA05831.1 ATLV01023045 KE525339 KFB48323.1 GBYB01003742 JAG73509.1 GDHC01003177 JAQ15452.1 GBHO01029251 JAG14353.1 GEDC01025447 JAS11851.1 KZ288226 PBC31884.1 JR047098 AEY60326.1 GAPW01001954 JAC11644.1 CVRI01000067 CRL06910.1 DS231833 EDS32298.1 AXCN02000879 AXCN02000880 APCN01001505 APCN01001506 AAAB01008960 EAA11525.4 QCYY01001875 ROT74611.1 KQ976423 KYM88771.1 GL888033 EGI69087.1 KQ983012 KYQ48355.1 GECZ01030087 GECZ01013471 JAS39682.1 JAS56298.1 GECU01015142 JAS92564.1 GECZ01019330 GECZ01006051 JAS50439.1 JAS63718.1
OWR43961.1 JTDY01000397 KOB77370.1 ODYU01002682 SOQ40495.1 NEVH01017000 PNF24697.1 PYGN01001983 PSN31927.1 KK852601 KDR20510.1 JRES01000714 KNC29023.1 CH477639 EAT37943.1 JXUM01089185 JXUM01089186 KQ563751 KXJ73378.1 CH902620 EDV31767.1 CM000361 CM002910 EDX03810.1 KMY88279.1 CH954177 EDV57849.1 CH480820 EDW54358.1 AE014134 AY051453 AAF52238.1 AAK92877.1 AGB92630.1 CM000157 EDW88940.1 OUUW01000004 SPP79537.1 AXCM01001573 AXCN02000839 CVRI01000037 CRK93608.1 CH933807 EDW12332.1 AAAB01008984 EAA15163.4 CH379060 KRT04300.1 JXJN01012999 EAL33883.3 CH479180 EDW29171.1 CH963920 EDW78156.1 CH940649 EDW64577.1 APCN01003745 AJVK01004152 AJVK01004153 AJVK01004154 CCAG010014402 CCAG010014403 CCAG010014404 DS231822 EDS25795.1 ADMH02000581 ETN65775.1 CP012523 ALC38287.1 GEDC01014208 JAS23090.1 CH916372 EDV98836.1 AJWK01031561 UFQT01000144 SSX20840.1 KK853243 KDR09457.1 AJWK01035509 AXCM01006480 AXCM01006481 GDAI01002816 JAI14787.1 APGK01014012 APGK01052024 KB741208 KB739171 KB631654 ENN72858.1 ENN82357.1 ERL85063.1 GEZM01077621 GEZM01077620 JAV63233.1 KQ971355 EFA05831.1 ATLV01023045 KE525339 KFB48323.1 GBYB01003742 JAG73509.1 GDHC01003177 JAQ15452.1 GBHO01029251 JAG14353.1 GEDC01025447 JAS11851.1 KZ288226 PBC31884.1 JR047098 AEY60326.1 GAPW01001954 JAC11644.1 CVRI01000067 CRL06910.1 DS231833 EDS32298.1 AXCN02000879 AXCN02000880 APCN01001505 APCN01001506 AAAB01008960 EAA11525.4 QCYY01001875 ROT74611.1 KQ976423 KYM88771.1 GL888033 EGI69087.1 KQ983012 KYQ48355.1 GECZ01030087 GECZ01013471 JAS39682.1 JAS56298.1 GECU01015142 JAS92564.1 GECZ01019330 GECZ01006051 JAS50439.1 JAS63718.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000235965
+ More
UP000245037 UP000027135 UP000037069 UP000008820 UP000069940 UP000249989 UP000007801 UP000192221 UP000076408 UP000000304 UP000008711 UP000001292 UP000075901 UP000000803 UP000002282 UP000268350 UP000075883 UP000075886 UP000183832 UP000009192 UP000007062 UP000075903 UP000075900 UP000001819 UP000075920 UP000092460 UP000075880 UP000092443 UP000008744 UP000075882 UP000075884 UP000075902 UP000091820 UP000076407 UP000007798 UP000008792 UP000075840 UP000092462 UP000092444 UP000002320 UP000000673 UP000092445 UP000092553 UP000069272 UP000078200 UP000001070 UP000092461 UP000095300 UP000095301 UP000192223 UP000019118 UP000030742 UP000007266 UP000030765 UP000242457 UP000283509 UP000078540 UP000007755 UP000075809
UP000245037 UP000027135 UP000037069 UP000008820 UP000069940 UP000249989 UP000007801 UP000192221 UP000076408 UP000000304 UP000008711 UP000001292 UP000075901 UP000000803 UP000002282 UP000268350 UP000075883 UP000075886 UP000183832 UP000009192 UP000007062 UP000075903 UP000075900 UP000001819 UP000075920 UP000092460 UP000075880 UP000092443 UP000008744 UP000075882 UP000075884 UP000075902 UP000091820 UP000076407 UP000007798 UP000008792 UP000075840 UP000092462 UP000092444 UP000002320 UP000000673 UP000092445 UP000092553 UP000069272 UP000078200 UP000001070 UP000092461 UP000095300 UP000095301 UP000192223 UP000019118 UP000030742 UP000007266 UP000030765 UP000242457 UP000283509 UP000078540 UP000007755 UP000075809
Pfam
PF03567 Sulfotransfer_2
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JKC2
A0A194PXB2
A0A194R0I1
A0A212ER84
A0A0L7LPL0
A0A2H1VI30
+ More
A0A2J7Q7Y6 A0A2P8XIU4 A0A067RLT5 A0A0L0C9C5 Q16TU6 A0A182GUH3 B3MLL8 A0A1W4UWA5 A0A182YFD2 B4Q3K6 B3N4U7 B4I1F1 A0A182SFU7 Q9VMS3 B4P075 A0A3B0JB11 A0A182MUA9 A0A182QLS7 A0A1J1HZT2 B4KI39 Q7PW21 A0A182V690 A0A182R9M9 A0A0R3NTN8 A0A182WH09 A0A1B0BES2 Q29M12 A0A182JML7 A0A1A9XRX3 B4G6Q7 A0A182KUN5 A0A182NEY8 A0A182U842 A0A1A9WUW0 A0A182WYR3 B4N0Z4 B4LRA7 A0A182I508 A0A1S4H0Z4 A0A1B0D8W1 A0A1B0FFN9 B0W1J7 W5JS76 A0A1A9ZK91 A0A0M4ENM4 A0A182F5F3 A0A1B6DBS3 A0A1A9VXY1 B4JPK6 A0A1B0CW14 A0A1I8PV94 A0A1I8MI65 A0A336LSE2 A0A067QKN1 A0A1B0CY71 A0A182S9P5 A0A182MSR1 A0A1W4WQX9 A0A182WKU9 A0A182YD58 A0A0K8TKD1 A0A182RIJ1 N6T5B1 A0A1Y1KP93 A0A182NIC2 D6WTL6 A0A084WDM9 A0A0C9QSZ1 A0A146M8T9 A0A0A9X699 A0A1B6CEK6 A0A182XMR2 A0A2A3EK66 A0A182IS71 V9IIL2 A0A182UV54 A0A182FE55 A0A023ETD2 A0A1J1J607 B0W401 A0A182QLH1 A0A182L309 A0A182HWM0 Q7Q6Q9 A0A3R7M829 A0A195BQC1 F4W9V1 A0A151WL04 A0A1B6EP42 A0A1B6J036 A0A1B6GMP7
A0A2J7Q7Y6 A0A2P8XIU4 A0A067RLT5 A0A0L0C9C5 Q16TU6 A0A182GUH3 B3MLL8 A0A1W4UWA5 A0A182YFD2 B4Q3K6 B3N4U7 B4I1F1 A0A182SFU7 Q9VMS3 B4P075 A0A3B0JB11 A0A182MUA9 A0A182QLS7 A0A1J1HZT2 B4KI39 Q7PW21 A0A182V690 A0A182R9M9 A0A0R3NTN8 A0A182WH09 A0A1B0BES2 Q29M12 A0A182JML7 A0A1A9XRX3 B4G6Q7 A0A182KUN5 A0A182NEY8 A0A182U842 A0A1A9WUW0 A0A182WYR3 B4N0Z4 B4LRA7 A0A182I508 A0A1S4H0Z4 A0A1B0D8W1 A0A1B0FFN9 B0W1J7 W5JS76 A0A1A9ZK91 A0A0M4ENM4 A0A182F5F3 A0A1B6DBS3 A0A1A9VXY1 B4JPK6 A0A1B0CW14 A0A1I8PV94 A0A1I8MI65 A0A336LSE2 A0A067QKN1 A0A1B0CY71 A0A182S9P5 A0A182MSR1 A0A1W4WQX9 A0A182WKU9 A0A182YD58 A0A0K8TKD1 A0A182RIJ1 N6T5B1 A0A1Y1KP93 A0A182NIC2 D6WTL6 A0A084WDM9 A0A0C9QSZ1 A0A146M8T9 A0A0A9X699 A0A1B6CEK6 A0A182XMR2 A0A2A3EK66 A0A182IS71 V9IIL2 A0A182UV54 A0A182FE55 A0A023ETD2 A0A1J1J607 B0W401 A0A182QLH1 A0A182L309 A0A182HWM0 Q7Q6Q9 A0A3R7M829 A0A195BQC1 F4W9V1 A0A151WL04 A0A1B6EP42 A0A1B6J036 A0A1B6GMP7
Ontologies
GO
PANTHER
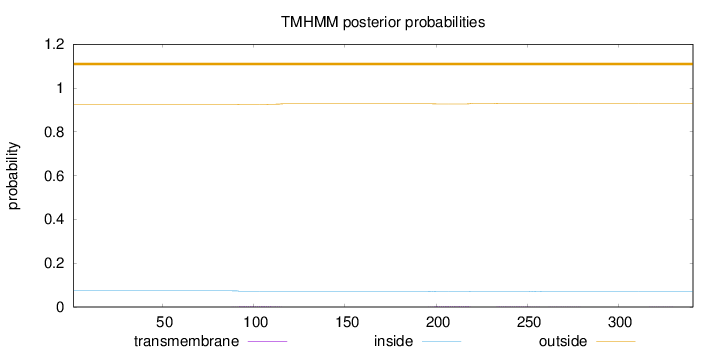
Topology
Subcellular location
Golgi apparatus membrane
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16307
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07440
outside
1 - 341
Population Genetic Test Statistics
Pi
262.91049
Theta
207.238498
Tajima's D
0.860973
CLR
0.252702
CSRT
0.614569271536423
Interpretation
Uncertain
