Gene
KWMTBOMO13232 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009976
Annotation
PREDICTED:_104_kDa_microneme/rhoptry_antigen_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.758
Sequence
CDS
ATGTATCAGAGACCACCGTTCTGGCGGGTGCCACACCAACGCGTGGACCCTGAAGACGAGCCCAGGAACTTTGGTCAACGAAAGGTGACGCCAGGTACTCCGGCCGGCAGGGACTTAGTCCGTGGGGATATCATCGCGAAGATCGACGATTATGACGCACGTGACCTTCGACACGAGGACGCACAGAATCTATTCAAGAATGCTCCAAATCAGATTAAATTAGTCATACAAAGGGGCGGTTCGAGTCCTCATCTATACTCTGTACCACGCACGACGGGCAGCTATGTAGGCCGTGTGCCGACACCCACATTTTATCACGAGCTCGCCGAGGACGTGGAACGCGAAGGACTGAGAGGTTCAGGCACGCCACTAGCGCCACGTTCGCCCATACCCAACGCAAGCGCCACACCATTCAGGACTCCAAGCCCGTTGCCTCCGTCATGGCGTGGTCCGGAATCTCTGCCTCGCAGCACACCGGTGCCACCTTCGCAATTCCAGCCACAACTGGACGGCGAATACAGAACCTATCTGCTGTCGCCTTCTTCCACGCGCACCGATGAGACCATCGATGAGTCTATTCAGAGGAACGACTGGAGCCGCAGTACGCGATCGCTCTCTGTTCGATACACGTCCATACCTACAAGTGTAACAATGTCGCTCAACCCCAACTTCTTCCCGAACGGATACCAGGATCCTAAACATCCTGAAGAAGAAGTTGTCAGCAACTGGCCGTACCGCACCACTCCTCTCGTGCTCCCGGGAGCTAAGGTCCGAAGGGAGCCTGGCCCCACAGAGAGTTACCTGCGTCATCACCCCAACCCAGCAATGAGGGCACCTCCTAACCACGACTACCGTGATACCCTTATGAAACAAAAGGTGGCAGAGTCGGTGCTGCAGCGAGTGGTTGGCGAAGAGGCACCTAAGGTGTTGCACAAGCAATTCAACTCTCCAATCAATCTATACTCGGAACAGAACATTGCAAACTCTATCAGGCAGCAAACTTCGCCTCTGCCAACTAACGGCCATTACGGACGGCCGCACGTTGTCAAGAGGCAATACGACCCCGCCAAGTCCGAGACGTACCGCGCGCTGCAGGAGGACGGCCTGCCCGACGCCGCCACCGAACTCTCCGCGCCCGTCGCCACCAAGGTCTTCACAGCGCCGACCAGCAAGAGGCCAGCGCCTACACCGAAACCGACGAAGCAGTCCGACGCGAAACCGAAAGGAAAACAAACGACCTTCGTAAATTCACTACACGAAGAACACATTCAACAGTCGAATTCATTCAAACGCCTCATGTTCAACGTACTTGGGGACACCGAATTCTAA
Protein
MYQRPPFWRVPHQRVDPEDEPRNFGQRKVTPGTPAGRDLVRGDIIAKIDDYDARDLRHEDAQNLFKNAPNQIKLVIQRGGSSPHLYSVPRTTGSYVGRVPTPTFYHELAEDVEREGLRGSGTPLAPRSPIPNASATPFRTPSPLPPSWRGPESLPRSTPVPPSQFQPQLDGEYRTYLLSPSSTRTDETIDESIQRNDWSRSTRSLSVRYTSIPTSVTMSLNPNFFPNGYQDPKHPEEEVVSNWPYRTTPLVLPGAKVRREPGPTESYLRHHPNPAMRAPPNHDYRDTLMKQKVAESVLQRVVGEEAPKVLHKQFNSPINLYSEQNIANSIRQQTSPLPTNGHYGRPHVVKRQYDPAKSETYRALQEDGLPDAATELSAPVATKVFTAPTSKRPAPTPKPTKQSDAKPKGKQTTFVNSLHEEHIQQSNSFKRLMFNVLGDTEF
Summary
Uniprot
A0A2H1V2M4
A0A3S2LFS1
I4DIK1
A0A0L7LPB7
A0A2J7RJJ5
A0A2J7RJJ3
+ More
A0A2J7RJJ9 A0A2J7RJJ7 A0A0Q9WHM8 A0A0Q9WI76 A0A151I117 A0A151XB97 A0A0Q9XBU0 A0A0R3P7K3 A0A3B0JAD3 A0A0Q9WRP5 A0A3B0J5U0 A0A0R3P980 A0A0Q9WZW3 A0A0Q9XKG2 A0A0R3P929 B4LEL6 A0A1W4US39 A0A195E804 A0A195F284 B4HK07 A0A0Q9XMT3 Q29D06 A0A0Q9WPB0 Q058U1 A0A0J9UHE3 A0A026WSY6 E1JI73 A0A0J9RRJ4 B4KZ65 A0A1W4UF88 A0A1W4UEP9 W8BZG7 A0A0A1XRR8 A0A0R1DUV9 Q95TZ7 A0A0J9RRZ2 A0A2J7RJJ8 A0A0Q9WP65 E2B6V5 Q8IQB4 A0A0J9RRM9 A0A0P8Y9N2 A0A1W4USC5 B3M679 A0A0Q9WZ91 A0A0P9C3B8 A0A034W4M8 A0A182QFX6 B4PFL1 W8BCA8 A0A0N8P0V3 A0A0M4EI34 T1PDC1 E2AHW2 A0A1J1I8S2 A0A034W0I2 A0A0M8ZNK4 A0A1J1ICE8 A0A1I8NBW7 A0A1I8NBW4 N6U6S5 A0A1J1I780 U4UGN0 A0A1I8Q0C5 A0A1I8Q081 A0A1I8Q0B0 A0A1I8Q0A1 A0A0Q9WIP2
A0A2J7RJJ9 A0A2J7RJJ7 A0A0Q9WHM8 A0A0Q9WI76 A0A151I117 A0A151XB97 A0A0Q9XBU0 A0A0R3P7K3 A0A3B0JAD3 A0A0Q9WRP5 A0A3B0J5U0 A0A0R3P980 A0A0Q9WZW3 A0A0Q9XKG2 A0A0R3P929 B4LEL6 A0A1W4US39 A0A195E804 A0A195F284 B4HK07 A0A0Q9XMT3 Q29D06 A0A0Q9WPB0 Q058U1 A0A0J9UHE3 A0A026WSY6 E1JI73 A0A0J9RRJ4 B4KZ65 A0A1W4UF88 A0A1W4UEP9 W8BZG7 A0A0A1XRR8 A0A0R1DUV9 Q95TZ7 A0A0J9RRZ2 A0A2J7RJJ8 A0A0Q9WP65 E2B6V5 Q8IQB4 A0A0J9RRM9 A0A0P8Y9N2 A0A1W4USC5 B3M679 A0A0Q9WZ91 A0A0P9C3B8 A0A034W4M8 A0A182QFX6 B4PFL1 W8BCA8 A0A0N8P0V3 A0A0M4EI34 T1PDC1 E2AHW2 A0A1J1I8S2 A0A034W0I2 A0A0M8ZNK4 A0A1J1ICE8 A0A1I8NBW7 A0A1I8NBW4 N6U6S5 A0A1J1I780 U4UGN0 A0A1I8Q0C5 A0A1I8Q081 A0A1I8Q0B0 A0A1I8Q0A1 A0A0Q9WIP2
Pubmed
EMBL
ODYU01000392
SOQ35097.1
RSAL01000018
RVE52832.1
AK401119
BAM17741.1
+ More
JTDY01000397 KOB77373.1 NEVH01002992 PNF41005.1 PNF41006.1 PNF41007.1 PNF41003.1 CH940647 KRF84173.1 KRF84179.1 KQ976580 KYM79892.1 KQ982335 KYQ57599.1 CH933809 KRG05805.1 CH379070 KRT09096.1 OUUW01000002 SPP77393.1 KRF84177.1 SPP77394.1 KRT09091.1 CH963847 KRF97736.1 KRG05812.1 KRT09093.1 EDW69101.2 KQ979568 KYN20949.1 KQ981864 KYN34199.1 CH480815 EDW40743.1 KRG05809.1 EAL30608.3 KRF97733.1 BT029127 AE014296 ABJ17060.1 ACZ94653.1 CM002912 KMY98435.1 KK107109 EZA59107.1 BT133470 ACZ94656.1 AFH97160.1 KMY98441.1 EDW17862.2 GAMC01011916 JAB94639.1 GBXI01000298 JAD13994.1 CM000159 KRK00966.1 AY058415 AAL13644.1 AAN11991.2 KMY98447.1 PNF41004.1 KRF97737.1 GL446009 EFN88596.1 AAN11990.2 KMY98438.1 CH902618 KPU78162.1 EDV39699.2 KRF97740.1 KPU78160.1 GAKP01009857 JAC49095.1 AXCN02000346 EDW93137.2 GAMC01011917 JAB94638.1 KPU78164.1 CP012525 ALC43451.1 KA646684 AFP61313.1 GL439602 EFN66952.1 CVRI01000043 CRK96114.1 GAKP01009851 JAC49101.1 KQ435969 KOX67895.1 CRK96113.1 APGK01037555 APGK01037556 APGK01037557 KB740948 ENN77350.1 CRK96112.1 KB632288 ERL91503.1 KRF84178.1
JTDY01000397 KOB77373.1 NEVH01002992 PNF41005.1 PNF41006.1 PNF41007.1 PNF41003.1 CH940647 KRF84173.1 KRF84179.1 KQ976580 KYM79892.1 KQ982335 KYQ57599.1 CH933809 KRG05805.1 CH379070 KRT09096.1 OUUW01000002 SPP77393.1 KRF84177.1 SPP77394.1 KRT09091.1 CH963847 KRF97736.1 KRG05812.1 KRT09093.1 EDW69101.2 KQ979568 KYN20949.1 KQ981864 KYN34199.1 CH480815 EDW40743.1 KRG05809.1 EAL30608.3 KRF97733.1 BT029127 AE014296 ABJ17060.1 ACZ94653.1 CM002912 KMY98435.1 KK107109 EZA59107.1 BT133470 ACZ94656.1 AFH97160.1 KMY98441.1 EDW17862.2 GAMC01011916 JAB94639.1 GBXI01000298 JAD13994.1 CM000159 KRK00966.1 AY058415 AAL13644.1 AAN11991.2 KMY98447.1 PNF41004.1 KRF97737.1 GL446009 EFN88596.1 AAN11990.2 KMY98438.1 CH902618 KPU78162.1 EDV39699.2 KRF97740.1 KPU78160.1 GAKP01009857 JAC49095.1 AXCN02000346 EDW93137.2 GAMC01011917 JAB94638.1 KPU78164.1 CP012525 ALC43451.1 KA646684 AFP61313.1 GL439602 EFN66952.1 CVRI01000043 CRK96114.1 GAKP01009851 JAC49101.1 KQ435969 KOX67895.1 CRK96113.1 APGK01037555 APGK01037556 APGK01037557 KB740948 ENN77350.1 CRK96112.1 KB632288 ERL91503.1 KRF84178.1
Proteomes
UP000283053
UP000037510
UP000235965
UP000008792
UP000078540
UP000075809
+ More
UP000009192 UP000001819 UP000268350 UP000007798 UP000192221 UP000078492 UP000078541 UP000001292 UP000000803 UP000053097 UP000002282 UP000008237 UP000007801 UP000075886 UP000092553 UP000000311 UP000183832 UP000053105 UP000095301 UP000019118 UP000030742 UP000095300
UP000009192 UP000001819 UP000268350 UP000007798 UP000192221 UP000078492 UP000078541 UP000001292 UP000000803 UP000053097 UP000002282 UP000008237 UP000007801 UP000075886 UP000092553 UP000000311 UP000183832 UP000053105 UP000095301 UP000019118 UP000030742 UP000095300
Interpro
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
A0A2H1V2M4
A0A3S2LFS1
I4DIK1
A0A0L7LPB7
A0A2J7RJJ5
A0A2J7RJJ3
+ More
A0A2J7RJJ9 A0A2J7RJJ7 A0A0Q9WHM8 A0A0Q9WI76 A0A151I117 A0A151XB97 A0A0Q9XBU0 A0A0R3P7K3 A0A3B0JAD3 A0A0Q9WRP5 A0A3B0J5U0 A0A0R3P980 A0A0Q9WZW3 A0A0Q9XKG2 A0A0R3P929 B4LEL6 A0A1W4US39 A0A195E804 A0A195F284 B4HK07 A0A0Q9XMT3 Q29D06 A0A0Q9WPB0 Q058U1 A0A0J9UHE3 A0A026WSY6 E1JI73 A0A0J9RRJ4 B4KZ65 A0A1W4UF88 A0A1W4UEP9 W8BZG7 A0A0A1XRR8 A0A0R1DUV9 Q95TZ7 A0A0J9RRZ2 A0A2J7RJJ8 A0A0Q9WP65 E2B6V5 Q8IQB4 A0A0J9RRM9 A0A0P8Y9N2 A0A1W4USC5 B3M679 A0A0Q9WZ91 A0A0P9C3B8 A0A034W4M8 A0A182QFX6 B4PFL1 W8BCA8 A0A0N8P0V3 A0A0M4EI34 T1PDC1 E2AHW2 A0A1J1I8S2 A0A034W0I2 A0A0M8ZNK4 A0A1J1ICE8 A0A1I8NBW7 A0A1I8NBW4 N6U6S5 A0A1J1I780 U4UGN0 A0A1I8Q0C5 A0A1I8Q081 A0A1I8Q0B0 A0A1I8Q0A1 A0A0Q9WIP2
A0A2J7RJJ9 A0A2J7RJJ7 A0A0Q9WHM8 A0A0Q9WI76 A0A151I117 A0A151XB97 A0A0Q9XBU0 A0A0R3P7K3 A0A3B0JAD3 A0A0Q9WRP5 A0A3B0J5U0 A0A0R3P980 A0A0Q9WZW3 A0A0Q9XKG2 A0A0R3P929 B4LEL6 A0A1W4US39 A0A195E804 A0A195F284 B4HK07 A0A0Q9XMT3 Q29D06 A0A0Q9WPB0 Q058U1 A0A0J9UHE3 A0A026WSY6 E1JI73 A0A0J9RRJ4 B4KZ65 A0A1W4UF88 A0A1W4UEP9 W8BZG7 A0A0A1XRR8 A0A0R1DUV9 Q95TZ7 A0A0J9RRZ2 A0A2J7RJJ8 A0A0Q9WP65 E2B6V5 Q8IQB4 A0A0J9RRM9 A0A0P8Y9N2 A0A1W4USC5 B3M679 A0A0Q9WZ91 A0A0P9C3B8 A0A034W4M8 A0A182QFX6 B4PFL1 W8BCA8 A0A0N8P0V3 A0A0M4EI34 T1PDC1 E2AHW2 A0A1J1I8S2 A0A034W0I2 A0A0M8ZNK4 A0A1J1ICE8 A0A1I8NBW7 A0A1I8NBW4 N6U6S5 A0A1J1I780 U4UGN0 A0A1I8Q0C5 A0A1I8Q081 A0A1I8Q0B0 A0A1I8Q0A1 A0A0Q9WIP2
PDB
1V5L
E-value=0.0711825,
Score=85
Ontologies
GO
Topology
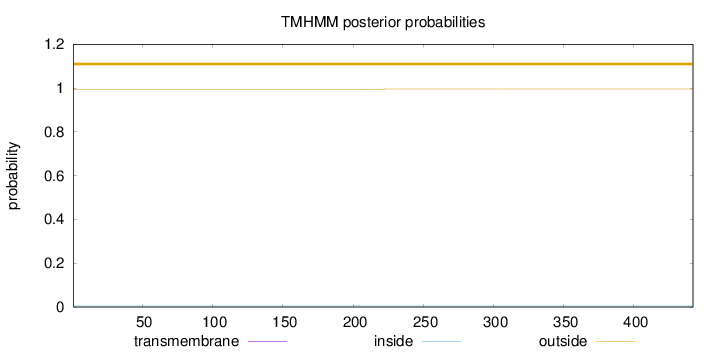
Length:
442
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00123
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00496
outside
1 - 442
Population Genetic Test Statistics
Pi
202.162458
Theta
175.536916
Tajima's D
-1.368216
CLR
1.579998
CSRT
0.0785460726963652
Interpretation
Possibly Positive selection
