Gene
KWMTBOMO13223
Pre Gene Modal
BGIBMGA009966
Annotation
PREDICTED:_D-2-hydroxyglutarate_dehydrogenase?_mitochondrial_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.717 Mitochondrial Reliability : 1.152
Sequence
CDS
ATGTTGAAATCAGTTGTGTTTTTATTGAGGTGTAATAGTAATAGAATCATTCGCGATCAAGTAAGGTATGCTTCACAAGCACTACCACAACTTTCTTCGGAAAAATACAACGTTAAGAGGAAACAATTTGGAACGGTTCAAGAGTCTGATATAAATTACTTTAAATCGATACTCAGCGAAGAAAGAGTGTTAACTGACGAAAATGATGTTTTGCCATTTAATATCGACTGGATTAAAAATTGCCGAGGGCAGTCAAAAGTTGTTTTAAAACCCAGAAGCACAGAGGAGGTTTCGAAAATTGTCAAGTATTGTAATGACAAACATTTGGCGGTATGTCCGCAAGGCGGGAACACTGGACTGGTGGGAGGCTCAGTGCCGGTCTTCGATGAAGTCGTTCTTTGTCTTGGTCTTCTGAACAAAATAATAAGTTTGGATGAAATATCTGGTGTTCTCATTTGTGAAGCGGGATGCATCCTTGAAAGCTTAGATACATGTGTCCGTGAGCGCAATCTAATAATGCCACTGGACTTAGGGGCAAAAGGTACTTGTCAAATTGGTGGTAATGCCAGCACCAATGCAGGAGGTCTGAGACTTCTCCGATACGGAAACTTACATGGTTCTATACTAGGCATTGAAGCGGTAAAAGCTGATGGCACCGTGATCGATTGTCTCACTACAATGAAGAAAGACAACACAGGATACCATTTAAAACATTTGTTTATCGGATCTGAGGGTACTCTAGGAATCGTCACTAAAGTTTCCATTAATTGCCCGGTTTTACCTAAAGCGGTCACGGTGGGATTCTTCGGGGTTAAAGATTTTCAAAGCGTCCTCAAACTGTACAAAAGCGCAAAACTTTCTTTAGGCGAAATATTGTCAGCATTTGAGATGGCCGATTTCAATGCGATCAATTCATCAGTTACGAATCTCAATTTACCGAACCCCATAGACGAATATCCCTTCTACGTGCTGGTCGAAACTCACGGTAGCGATGAAGGTCACGACGCCGAAAAATTAAATCGATTCCTTGATAAAGAAATGGAAAGTGGACTAATTATCGACGGTACTGTAACATCCGAACCCGCCAAAATGCAGACCATCTGGAACCTTCGAGAGAGTATAGCCGGCTCGAGTCTAACCCGAGGCTACGTTTTTAAATACGACGTGTCTATACCACTCGGAAATTATTATGATTTAGTTCCAATGCTACGAAAGAAACTGGGCAATAAGGCCCTCGATGTATACGGATATGGACATATAGGGGATAGCAATATTCATATAAACATCGTAGTACCGGAATATACGAAAGAAATAGGCTCTCAAATAGAACCTTATGTGTTTGAAGAAGTGAGCAAATTGAACGGGTCGATTAGCGCAGAGCACGGCGTCGGTTTTCGCAAGCCACAGTACATTCATTACAGTAAAAATGAATCTGCCCTTGATCTCATGAGGGAACTGAAGACGCTTATGGACCCGAAGGGAATATTGAACCCATATAAAGTTCTACCTGAGAAATAA
Protein
MLKSVVFLLRCNSNRIIRDQVRYASQALPQLSSEKYNVKRKQFGTVQESDINYFKSILSEERVLTDENDVLPFNIDWIKNCRGQSKVVLKPRSTEEVSKIVKYCNDKHLAVCPQGGNTGLVGGSVPVFDEVVLCLGLLNKIISLDEISGVLICEAGCILESLDTCVRERNLIMPLDLGAKGTCQIGGNASTNAGGLRLLRYGNLHGSILGIEAVKADGTVIDCLTTMKKDNTGYHLKHLFIGSEGTLGIVTKVSINCPVLPKAVTVGFFGVKDFQSVLKLYKSAKLSLGEILSAFEMADFNAINSSVTNLNLPNPIDEYPFYVLVETHGSDEGHDAEKLNRFLDKEMESGLIIDGTVTSEPAKMQTIWNLRESIAGSSLTRGYVFKYDVSIPLGNYYDLVPMLRKKLGNKALDVYGYGHIGDSNIHINIVVPEYTKEIGSQIEPYVFEEVSKLNGSISAEHGVGFRKPQYIHYSKNESALDLMRELKTLMDPKGILNPYKVLPEK
Summary
Uniprot
H9JKB5
A0A2A4J6D8
A0A194R231
A0A2H1V529
A0A194PX94
A0A3S2LSC6
+ More
A0A212EM57 A0A1Y1L3M7 A0A2J7QG50 A0A2J7QG54 A0A1B6DRW9 A0A1B6CTR9 A0A067QYH9 D6WE94 A0A2M4AHA9 A0A2M4AHF7 A0A182FSC3 A0A1Q3FJQ0 B0W312 W5J8Q1 K7J9X2 A0A232F3B9 A0A182H4A8 Q178E1 A0A1B0GIE0 A0A1L8E0B9 A0A1L8E0H0 A0A182P6Q8 A0A182NLE3 A0A084VNS6 A0A1A9X1G3 A0A182UN40 A0A182QXF0 A0A182W1F7 A0A182MKB9 A0A182I7Q2 A0A182XJ58 A0A182L792 Q7QB44 A0A182ISZ3 A0A182RYR0 A0A1B6H6E0 A0A1B6E3S3 A0A1B6C7D9 A0A1J1HQ30 A0A1B0AEB3 A0A1I8P1Z4 A0A0L7RBM8 A0A1B6H1T9 C3ZLK3 A0A336KFA2 V4BQX2 D3TMR5 A0A1A9UL97 A0A0A1WLC2 E2C5C8 A0A195D861 A0A158NRY9 A0A226EPN0 A0A195BPW6 A0A0L0C422 A0A1A9YNU9 F4WQQ2 A0A1B0APM5 D6WE92 A0A034VBG7 A0A1W4WY92 A0A0K8WEH3 A0A034VTF1 H3A1S5 A0A1I8NGT2 A0A0K8W8C2 A0A1S3IE10 A0A1S3IG95 A0A1B6FGW0 A0A3L8D3F0 A0A1S3IEB0 A0A1S3IF60 A0A026W6H1 A0A210R5A8 A0A0N0BG86 G1K8L8 E4Z4C4 A0A1V4KQU7 A0A0K8RU31 A0A1D2MZC9 H0Z684 F6PH88 A0A1Y1M291 A0A1W2WPB0 A0A3P8PF93 H0Z689 G1Q363 S7MGC0 A0A3B4GCP3 A0A3P9ATN6 F6W0L1 A0A218UBZ6 A0A3Q2V2A8 A0A1U7SFV0
A0A212EM57 A0A1Y1L3M7 A0A2J7QG50 A0A2J7QG54 A0A1B6DRW9 A0A1B6CTR9 A0A067QYH9 D6WE94 A0A2M4AHA9 A0A2M4AHF7 A0A182FSC3 A0A1Q3FJQ0 B0W312 W5J8Q1 K7J9X2 A0A232F3B9 A0A182H4A8 Q178E1 A0A1B0GIE0 A0A1L8E0B9 A0A1L8E0H0 A0A182P6Q8 A0A182NLE3 A0A084VNS6 A0A1A9X1G3 A0A182UN40 A0A182QXF0 A0A182W1F7 A0A182MKB9 A0A182I7Q2 A0A182XJ58 A0A182L792 Q7QB44 A0A182ISZ3 A0A182RYR0 A0A1B6H6E0 A0A1B6E3S3 A0A1B6C7D9 A0A1J1HQ30 A0A1B0AEB3 A0A1I8P1Z4 A0A0L7RBM8 A0A1B6H1T9 C3ZLK3 A0A336KFA2 V4BQX2 D3TMR5 A0A1A9UL97 A0A0A1WLC2 E2C5C8 A0A195D861 A0A158NRY9 A0A226EPN0 A0A195BPW6 A0A0L0C422 A0A1A9YNU9 F4WQQ2 A0A1B0APM5 D6WE92 A0A034VBG7 A0A1W4WY92 A0A0K8WEH3 A0A034VTF1 H3A1S5 A0A1I8NGT2 A0A0K8W8C2 A0A1S3IE10 A0A1S3IG95 A0A1B6FGW0 A0A3L8D3F0 A0A1S3IEB0 A0A1S3IF60 A0A026W6H1 A0A210R5A8 A0A0N0BG86 G1K8L8 E4Z4C4 A0A1V4KQU7 A0A0K8RU31 A0A1D2MZC9 H0Z684 F6PH88 A0A1Y1M291 A0A1W2WPB0 A0A3P8PF93 H0Z689 G1Q363 S7MGC0 A0A3B4GCP3 A0A3P9ATN6 F6W0L1 A0A218UBZ6 A0A3Q2V2A8 A0A1U7SFV0
Pubmed
19121390
26354079
22118469
28004739
24845553
18362917
+ More
19820115 20920257 23761445 20075255 28648823 26483478 17510324 24438588 20966253 12364791 14747013 17210077 18563158 23254933 20353571 25830018 20798317 21347285 26108605 21719571 25348373 9215903 25315136 30249741 24508170 28812685 21097902 25727380 27289101 20360741 17495919 21993624 25186727 12481130 15114417
19820115 20920257 23761445 20075255 28648823 26483478 17510324 24438588 20966253 12364791 14747013 17210077 18563158 23254933 20353571 25830018 20798317 21347285 26108605 21719571 25348373 9215903 25315136 30249741 24508170 28812685 21097902 25727380 27289101 20360741 17495919 21993624 25186727 12481130 15114417
EMBL
BABH01036767
NWSH01002706
PCG67645.1
KQ460883
KPJ11310.1
ODYU01000696
+ More
SOQ35899.1 KQ459586 KPI97971.1 RSAL01000018 RVE52823.1 AGBW02013899 OWR42580.1 GEZM01066100 JAV68184.1 NEVH01014835 PNF27569.1 PNF27570.1 GEDC01008925 JAS28373.1 GEDC01020675 GEDC01007325 JAS16623.1 JAS29973.1 KK853191 KDR10015.1 KQ971326 EEZ99904.2 GGFK01006843 MBW40164.1 GGFK01006856 MBW40177.1 GFDL01007194 JAV27851.1 DS231830 EDS30720.1 ADMH02001952 ETN60341.1 AAZX01018770 NNAY01001148 OXU24980.1 JXUM01109092 KQ565289 KXJ71128.1 CH477365 EAT42562.1 AJWK01013775 GFDF01001904 JAV12180.1 GFDF01001905 JAV12179.1 ATLV01014857 ATLV01014858 KE524991 KFB39620.1 AXCN02000130 AXCM01012580 APCN01002523 APCN01002524 AAAB01008880 EAA08571.4 GECU01037475 JAS70231.1 GEDC01004737 JAS32561.1 GEDC01028138 JAS09160.1 CVRI01000009 CRK88582.1 KQ414617 KOC68161.1 GECZ01018543 GECZ01014147 GECZ01001161 JAS51226.1 JAS55622.1 JAS68608.1 GG666641 EEN46640.1 UFQS01000108 UFQT01000108 SSW99730.1 SSX20110.1 KB202283 ESO91314.1 CCAG010004528 EZ422717 ADD18993.1 GBXI01014625 JAC99666.1 GL452770 EFN76861.1 KQ981153 KYN09061.1 ADTU01024334 LNIX01000002 OXA59575.1 KQ976424 KYM88562.1 JRES01000940 KNC27030.1 GL888275 EGI63415.1 JXJN01001483 EFA01450.2 GAKP01019103 JAC39849.1 GDHF01002870 JAI49444.1 GAKP01013847 JAC45105.1 AFYH01255629 AFYH01255630 GDHF01004928 JAI47386.1 GECZ01020354 JAS49415.1 QOIP01000014 RLU14766.1 KK107405 EZA51226.1 NEDP02000262 OWF56209.1 KQ435793 KOX74198.1 FN657273 CBY42552.1 LSYS01002180 OPJ86783.1 GBKC01001669 JAG44401.1 LJIJ01000375 ODM98174.1 ABQF01030504 GEZM01042829 JAV79561.1 AAPE02004541 AAPE02004542 KE161207 EPQ02370.1 EAAA01000360 MUZQ01000464 OWK51111.1
SOQ35899.1 KQ459586 KPI97971.1 RSAL01000018 RVE52823.1 AGBW02013899 OWR42580.1 GEZM01066100 JAV68184.1 NEVH01014835 PNF27569.1 PNF27570.1 GEDC01008925 JAS28373.1 GEDC01020675 GEDC01007325 JAS16623.1 JAS29973.1 KK853191 KDR10015.1 KQ971326 EEZ99904.2 GGFK01006843 MBW40164.1 GGFK01006856 MBW40177.1 GFDL01007194 JAV27851.1 DS231830 EDS30720.1 ADMH02001952 ETN60341.1 AAZX01018770 NNAY01001148 OXU24980.1 JXUM01109092 KQ565289 KXJ71128.1 CH477365 EAT42562.1 AJWK01013775 GFDF01001904 JAV12180.1 GFDF01001905 JAV12179.1 ATLV01014857 ATLV01014858 KE524991 KFB39620.1 AXCN02000130 AXCM01012580 APCN01002523 APCN01002524 AAAB01008880 EAA08571.4 GECU01037475 JAS70231.1 GEDC01004737 JAS32561.1 GEDC01028138 JAS09160.1 CVRI01000009 CRK88582.1 KQ414617 KOC68161.1 GECZ01018543 GECZ01014147 GECZ01001161 JAS51226.1 JAS55622.1 JAS68608.1 GG666641 EEN46640.1 UFQS01000108 UFQT01000108 SSW99730.1 SSX20110.1 KB202283 ESO91314.1 CCAG010004528 EZ422717 ADD18993.1 GBXI01014625 JAC99666.1 GL452770 EFN76861.1 KQ981153 KYN09061.1 ADTU01024334 LNIX01000002 OXA59575.1 KQ976424 KYM88562.1 JRES01000940 KNC27030.1 GL888275 EGI63415.1 JXJN01001483 EFA01450.2 GAKP01019103 JAC39849.1 GDHF01002870 JAI49444.1 GAKP01013847 JAC45105.1 AFYH01255629 AFYH01255630 GDHF01004928 JAI47386.1 GECZ01020354 JAS49415.1 QOIP01000014 RLU14766.1 KK107405 EZA51226.1 NEDP02000262 OWF56209.1 KQ435793 KOX74198.1 FN657273 CBY42552.1 LSYS01002180 OPJ86783.1 GBKC01001669 JAG44401.1 LJIJ01000375 ODM98174.1 ABQF01030504 GEZM01042829 JAV79561.1 AAPE02004541 AAPE02004542 KE161207 EPQ02370.1 EAAA01000360 MUZQ01000464 OWK51111.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000235965 UP000027135 UP000007266 UP000069272 UP000002320 UP000000673 UP000002358 UP000215335 UP000069940 UP000249989 UP000008820 UP000092461 UP000075885 UP000075884 UP000030765 UP000091820 UP000075903 UP000075886 UP000075920 UP000075883 UP000075840 UP000076407 UP000075882 UP000007062 UP000075880 UP000075900 UP000183832 UP000092445 UP000095300 UP000053825 UP000001554 UP000030746 UP000092444 UP000078200 UP000008237 UP000078492 UP000005205 UP000198287 UP000078540 UP000037069 UP000092443 UP000007755 UP000092460 UP000192223 UP000008672 UP000095301 UP000085678 UP000279307 UP000053097 UP000242188 UP000053105 UP000001646 UP000190648 UP000094527 UP000007754 UP000002280 UP000265100 UP000001074 UP000261460 UP000265160 UP000008144 UP000197619 UP000264840 UP000189705
UP000235965 UP000027135 UP000007266 UP000069272 UP000002320 UP000000673 UP000002358 UP000215335 UP000069940 UP000249989 UP000008820 UP000092461 UP000075885 UP000075884 UP000030765 UP000091820 UP000075903 UP000075886 UP000075920 UP000075883 UP000075840 UP000076407 UP000075882 UP000007062 UP000075880 UP000075900 UP000183832 UP000092445 UP000095300 UP000053825 UP000001554 UP000030746 UP000092444 UP000078200 UP000008237 UP000078492 UP000005205 UP000198287 UP000078540 UP000037069 UP000092443 UP000007755 UP000092460 UP000192223 UP000008672 UP000095301 UP000085678 UP000279307 UP000053097 UP000242188 UP000053105 UP000001646 UP000190648 UP000094527 UP000007754 UP000002280 UP000265100 UP000001074 UP000261460 UP000265160 UP000008144 UP000197619 UP000264840 UP000189705
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JKB5
A0A2A4J6D8
A0A194R231
A0A2H1V529
A0A194PX94
A0A3S2LSC6
+ More
A0A212EM57 A0A1Y1L3M7 A0A2J7QG50 A0A2J7QG54 A0A1B6DRW9 A0A1B6CTR9 A0A067QYH9 D6WE94 A0A2M4AHA9 A0A2M4AHF7 A0A182FSC3 A0A1Q3FJQ0 B0W312 W5J8Q1 K7J9X2 A0A232F3B9 A0A182H4A8 Q178E1 A0A1B0GIE0 A0A1L8E0B9 A0A1L8E0H0 A0A182P6Q8 A0A182NLE3 A0A084VNS6 A0A1A9X1G3 A0A182UN40 A0A182QXF0 A0A182W1F7 A0A182MKB9 A0A182I7Q2 A0A182XJ58 A0A182L792 Q7QB44 A0A182ISZ3 A0A182RYR0 A0A1B6H6E0 A0A1B6E3S3 A0A1B6C7D9 A0A1J1HQ30 A0A1B0AEB3 A0A1I8P1Z4 A0A0L7RBM8 A0A1B6H1T9 C3ZLK3 A0A336KFA2 V4BQX2 D3TMR5 A0A1A9UL97 A0A0A1WLC2 E2C5C8 A0A195D861 A0A158NRY9 A0A226EPN0 A0A195BPW6 A0A0L0C422 A0A1A9YNU9 F4WQQ2 A0A1B0APM5 D6WE92 A0A034VBG7 A0A1W4WY92 A0A0K8WEH3 A0A034VTF1 H3A1S5 A0A1I8NGT2 A0A0K8W8C2 A0A1S3IE10 A0A1S3IG95 A0A1B6FGW0 A0A3L8D3F0 A0A1S3IEB0 A0A1S3IF60 A0A026W6H1 A0A210R5A8 A0A0N0BG86 G1K8L8 E4Z4C4 A0A1V4KQU7 A0A0K8RU31 A0A1D2MZC9 H0Z684 F6PH88 A0A1Y1M291 A0A1W2WPB0 A0A3P8PF93 H0Z689 G1Q363 S7MGC0 A0A3B4GCP3 A0A3P9ATN6 F6W0L1 A0A218UBZ6 A0A3Q2V2A8 A0A1U7SFV0
A0A212EM57 A0A1Y1L3M7 A0A2J7QG50 A0A2J7QG54 A0A1B6DRW9 A0A1B6CTR9 A0A067QYH9 D6WE94 A0A2M4AHA9 A0A2M4AHF7 A0A182FSC3 A0A1Q3FJQ0 B0W312 W5J8Q1 K7J9X2 A0A232F3B9 A0A182H4A8 Q178E1 A0A1B0GIE0 A0A1L8E0B9 A0A1L8E0H0 A0A182P6Q8 A0A182NLE3 A0A084VNS6 A0A1A9X1G3 A0A182UN40 A0A182QXF0 A0A182W1F7 A0A182MKB9 A0A182I7Q2 A0A182XJ58 A0A182L792 Q7QB44 A0A182ISZ3 A0A182RYR0 A0A1B6H6E0 A0A1B6E3S3 A0A1B6C7D9 A0A1J1HQ30 A0A1B0AEB3 A0A1I8P1Z4 A0A0L7RBM8 A0A1B6H1T9 C3ZLK3 A0A336KFA2 V4BQX2 D3TMR5 A0A1A9UL97 A0A0A1WLC2 E2C5C8 A0A195D861 A0A158NRY9 A0A226EPN0 A0A195BPW6 A0A0L0C422 A0A1A9YNU9 F4WQQ2 A0A1B0APM5 D6WE92 A0A034VBG7 A0A1W4WY92 A0A0K8WEH3 A0A034VTF1 H3A1S5 A0A1I8NGT2 A0A0K8W8C2 A0A1S3IE10 A0A1S3IG95 A0A1B6FGW0 A0A3L8D3F0 A0A1S3IEB0 A0A1S3IF60 A0A026W6H1 A0A210R5A8 A0A0N0BG86 G1K8L8 E4Z4C4 A0A1V4KQU7 A0A0K8RU31 A0A1D2MZC9 H0Z684 F6PH88 A0A1Y1M291 A0A1W2WPB0 A0A3P8PF93 H0Z689 G1Q363 S7MGC0 A0A3B4GCP3 A0A3P9ATN6 F6W0L1 A0A218UBZ6 A0A3Q2V2A8 A0A1U7SFV0
PDB
3PM9
E-value=1.67794e-81,
Score=772
Ontologies
GO
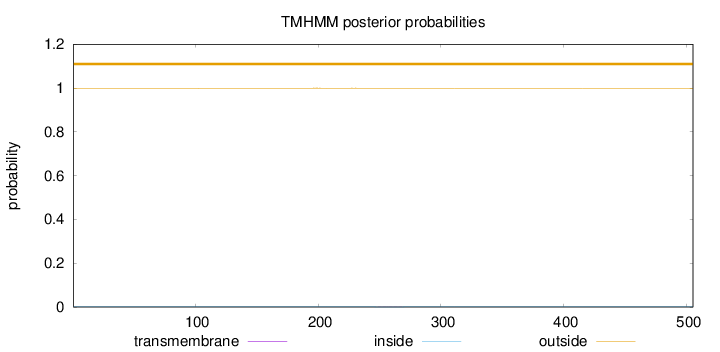
Topology
Length:
505
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00565999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00066
outside
1 - 505
Population Genetic Test Statistics
Pi
236.23898
Theta
175.888312
Tajima's D
1.063229
CLR
0.159813
CSRT
0.678866056697165
Interpretation
Uncertain
