Pre Gene Modal
BGIBMGA009985
Annotation
PREDICTED:_BRCA1-A_complex_subunit_BRE-like_[Amyelois_transitella]
Full name
BRISC and BRCA1-A complex member 2
Alternative Name
BRCA1-A complex subunit BRE
BRCA1/BRCA2-containing complex subunit 45
Brain and reproductive organ-expressed protein
BRCA1/BRCA2-containing complex subunit 45
Brain and reproductive organ-expressed protein
Location in the cell
Cytoplasmic Reliability : 1.925
Sequence
CDS
ATGGGAACTGAAGCTTTTGGATATTTCAAAAATTTATCTCCAGCATTTCGTCCTTACGTACAAACATTATGTGAAGAATTAAAACTTGGCCTCTGCAAGACTAAAATAGATGTTGAAAGAATATCTTGTTTACCTGATGGAGAGGAATGCCAATTCCGTCTGCTATTACCTTATTGTTCTAAGAAATTGAAATGGGAAATATTATTTGATGTATCAATACCATGGTTTGCTCCGGACTTTAAGTTTGATGATGAAAGTTTTTTGATAAGTGAGGATGAAAACTTTCTTGAAGAGAAAGTCCCAAGTCTTGCTAAATGGAATGAAAGTGATCCTAGAGCTTTGAGTAATGTGATATTTGAGTTGGTCAATCTATACAAAAGTCATCAGATTAGAAAACTGAGTGAGGATGATTCTTCAAGGGCTTATTTTGAATACAGTGCATTACTTGGTGATTCACTCATCTCTGAAAGTAACATCGAAGTTTGGGTTGGAACACATGTCGTTGAATTTTTGATAAAACTTAATGTTGACGTTGGAAGATTATCAGAGCTTTATAGTGATGGCATTGAAGAGAATCCCGGCATTGACACAGCATTGTTACTAGTGAGATATCCAAACTCGACAAACGCAGAACTGATACTATCACCTCTTCTTAATAAAGCCCTGGGGAACATTTCATTACCGCAAATGCATCCATCTACTGTTCTTATGGATTATGTACCTATGGTTACCGAACTGCTGAACAGAAAGATACATGATGTACTTGTCAATGAAAAATTGAAAAGGGATTTATTAGCATTTTTGGTTGTTAAATATGAAGGTGCCATATTGGAACACGATATAAACAGTGCTGCCTTTCTATTTGAAATCAATGACTTCCATTGGATTTTAACAGTAGAATTGGGTCAATTTCCAGAAAAATCTCCAATTTTGACCTTAAGATCAGTTTACCACAGCAGTAAAGGAAAACCAAAATACAAAGTTCTCTCTTCTTGTCCATTAAACAACTTTGAAGAATATGTGGAACAACAAGTAGAATTATTCAAAAACGAATGTAAAGGATCAGTAACTAGTAGTCAACTCGTAAGCTTGGAAAATTAA
Protein
MGTEAFGYFKNLSPAFRPYVQTLCEELKLGLCKTKIDVERISCLPDGEECQFRLLLPYCSKKLKWEILFDVSIPWFAPDFKFDDESFLISEDENFLEEKVPSLAKWNESDPRALSNVIFELVNLYKSHQIRKLSEDDSSRAYFEYSALLGDSLISESNIEVWVGTHVVEFLIKLNVDVGRLSELYSDGIEENPGIDTALLLVRYPNSTNAELILSPLLNKALGNISLPQMHPSTVLMDYVPMVTELLNRKIHDVLVNEKLKRDLLAFLVVKYEGAILEHDINSAAFLFEINDFHWILTVELGQFPEKSPILTLRSVYHSSKGKPKYKVLSSCPLNNFEEYVEQQVELFKNECKGSVTSSQLVSLEN
Summary
Description
Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the BRCA1-BARD1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX. In the BRCA1-A complex, it acts as an adapter that bridges the interaction between BABAM1/NBA1 and the rest of the complex, thereby being required for the complex integrity and modulating the E3 ubiquitin ligase activity of the BRCA1-BARD1 heterodimer. Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates. Within the BRISC complex, acts as an adapter that bridges the interaction between BABAM1/NBA1 and the rest of the complex, thereby being required for the complex integrity. The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating NUMA1. The BRISC complex plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor IFNAR1; deubiquitination increases IFNAR1 activity by enhancing its stability and cell surface expression. Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in IFNAR1 deubiquitination. May play a role in homeostasis or cellular differentiation in cells of neural, epithelial and germline origins. May also act as a death receptor-associated anti-apoptotic protein, which inhibits the mitochondrial apoptotic pathway. May regulate TNF-alpha signaling through its interactions with TNFRSF1A; however these effects may be indirect.
Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the brca1-bard1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX. In the BRCA1-A complex, it acts as an adapter that bridges the interaction between babam1/nba1 and the rest of the complex, thereby being required for the complex integrity and modulating the E3 ubiquitin ligase activity of the brca1-bard1 heterodimer. Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates. Within the BRISC complex, acts as an adapter that bridges the interaction between babam1/nba1 and the rest of the complex, thereby being required for the complex integrity. The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating numa1. The BRISC complex plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor ifnar1; deubiquitination increases ifnar1 activity by enhancing its stability and cell surface expression. Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in ifnar1 deubiquitination. May play a role in homeostasis or cellular differentiation in cells of neural, epithelial and germline origins. May also act as a death receptor-associated anti-apoptotic protein, which inhibits the mitochondrial apoptotic pathway.
Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the BRCA1-BARD1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX (PubMed:17525341, PubMed:19261746, PubMed:19261749, PubMed:19261748). In the BRCA1-A complex, it acts as an adapter that bridges the interaction between BABAM1/NBA1 and the rest of the complex, thereby being required for the complex integrity and modulating the E3 ubiquitin ligase activity of the BRCA1-BARD1 heterodimer (PubMed:21282113, PubMed:19261748). Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates (PubMed:19214193, PubMed:24075985, PubMed:25283148, PubMed:26195665). Within the BRISC complex, acts as an adapter that bridges the interaction between BABAM1/NBA1 and the rest of the complex, thereby being required for the complex integrity (PubMed:21282113). The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating NUMA1 (PubMed:26195665). The BRISC complex plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor IFNAR1; deubiquitination increases IFNAR1 activity by enhancing its stability and cell surface expression (PubMed:24075985). Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in IFNAR1 deubiquitination (PubMed:24075985). May play a role in homeostasis or cellular differentiation in cells of neural, epithelial and germline origins. May also act as a death receptor-associated anti-apoptotic protein, which inhibits the mitochondrial apoptotic pathway. May regulate TNF-alpha signaling through its interactions with TNFRSF1A; however these effects may be indirect (PubMed:15465831).
Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the brca1-bard1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX. In the BRCA1-A complex, it acts as an adapter that bridges the interaction between babam1/nba1 and the rest of the complex, thereby being required for the complex integrity and modulating the E3 ubiquitin ligase activity of the brca1-bard1 heterodimer. Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates. Within the BRISC complex, acts as an adapter that bridges the interaction between babam1/nba1 and the rest of the complex, thereby being required for the complex integrity. The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating numa1. The BRISC complex plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor ifnar1; deubiquitination increases ifnar1 activity by enhancing its stability and cell surface expression. Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in ifnar1 deubiquitination. May play a role in homeostasis or cellular differentiation in cells of neural, epithelial and germline origins. May also act as a death receptor-associated anti-apoptotic protein, which inhibits the mitochondrial apoptotic pathway.
Component of the BRCA1-A complex, a complex that specifically recognizes 'Lys-63'-linked ubiquitinated histones H2A and H2AX at DNA lesions sites, leading to target the BRCA1-BARD1 heterodimer to sites of DNA damage at double-strand breaks (DSBs). The BRCA1-A complex also possesses deubiquitinase activity that specifically removes 'Lys-63'-linked ubiquitin on histones H2A and H2AX (PubMed:17525341, PubMed:19261746, PubMed:19261749, PubMed:19261748). In the BRCA1-A complex, it acts as an adapter that bridges the interaction between BABAM1/NBA1 and the rest of the complex, thereby being required for the complex integrity and modulating the E3 ubiquitin ligase activity of the BRCA1-BARD1 heterodimer (PubMed:21282113, PubMed:19261748). Component of the BRISC complex, a multiprotein complex that specifically cleaves 'Lys-63'-linked ubiquitin in various substrates (PubMed:19214193, PubMed:24075985, PubMed:25283148, PubMed:26195665). Within the BRISC complex, acts as an adapter that bridges the interaction between BABAM1/NBA1 and the rest of the complex, thereby being required for the complex integrity (PubMed:21282113). The BRISC complex is required for normal mitotic spindle assembly and microtubule attachment to kinetochores via its role in deubiquitinating NUMA1 (PubMed:26195665). The BRISC complex plays a role in interferon signaling via its role in the deubiquitination of the interferon receptor IFNAR1; deubiquitination increases IFNAR1 activity by enhancing its stability and cell surface expression (PubMed:24075985). Down-regulates the response to bacterial lipopolysaccharide (LPS) via its role in IFNAR1 deubiquitination (PubMed:24075985). May play a role in homeostasis or cellular differentiation in cells of neural, epithelial and germline origins. May also act as a death receptor-associated anti-apoptotic protein, which inhibits the mitochondrial apoptotic pathway. May regulate TNF-alpha signaling through its interactions with TNFRSF1A; however these effects may be indirect (PubMed:15465831).
Subunit
Component of the ARISC complex, at least composed of UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. In the BRCA1-A complex, interacts directly with ABRAXAS1, BRCC3/BRCC36 and BABAM1/NBA1. Binds polyubiquitin. Component of the BRISC complex, at least composed of ABRAXAS2, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1.
Component of the ARISC complex, at least composed of uimc1/rap80, abraxas1, brcc3/brcc36, babam2 and babam1/nba1. Component of the BRCA1-A complex, at least composed of brca1, bard1, uimc1/rap80, abraxas1, brcc3/brcc36, babam2 and babam1/nba1. In the BRCA1-A complex, interacts directly with abraxas1, brcc3/brcc36 and babam1/nba1. Binds polyubiquitin. Component of the BRISC complex, at least composed of abraxas2, brcc3/brcc36, babam2 and babam1/nba1.
Component of the ARISC complex, at least composed of UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. In the BRCA1-A complex, interacts directly with ABRAXAS1, BRCC3/BRCC36 and BABAM1/NBA1. Binds polyubiquitin. Component of the BRISC complex, at least composed of ABRAXAS2, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. Identified in a complex with SHMT2 and the other subunits of the BRISC complex. Component of the BRCA1/BRCA2 containing complex (BRCC), which also contains BRCA1, BRCA2, BARD1, BRCC3/BRCC36 and RAD51. BRCC is a ubiquitin E3 ligase complex that enhances cellular survival following DNA damage. May interact with FAS and TNFRSF1A.
Component of the ARISC complex, at least composed of UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:21282113, PubMed:24075985). Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. In the BRCA1-A complex, interacts directly with ABRAXAS1, BRCC3/BRCC36 and BABAM1/NBA1. Binds polyubiquitin. Component of the BRISC complex, at least composed of ABRAXAS2, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:19214193, PubMed:21282113, PubMed:24075985, PubMed:25283148). Identified in a complex with SHMT2 and the other subunits of the BRISC complex (PubMed:24075985). Component of the BRCA1/BRCA2 containing complex (BRCC), which also contains BRCA1, BRCA2, BARD1, BRCC3/BRCC36 and RAD51. BRCC is a ubiquitin E3 ligase complex that enhances cellular survival following DNA damage. May interact with FAS and TNFRSF1A (PubMed:15465831).
Component of the ARISC complex, at least composed of uimc1/rap80, abraxas1, brcc3/brcc36, babam2 and babam1/nba1. Component of the BRCA1-A complex, at least composed of brca1, bard1, uimc1/rap80, abraxas1, brcc3/brcc36, babam2 and babam1/nba1. In the BRCA1-A complex, interacts directly with abraxas1, brcc3/brcc36 and babam1/nba1. Binds polyubiquitin. Component of the BRISC complex, at least composed of abraxas2, brcc3/brcc36, babam2 and babam1/nba1.
Component of the ARISC complex, at least composed of UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. In the BRCA1-A complex, interacts directly with ABRAXAS1, BRCC3/BRCC36 and BABAM1/NBA1. Binds polyubiquitin. Component of the BRISC complex, at least composed of ABRAXAS2, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. Identified in a complex with SHMT2 and the other subunits of the BRISC complex. Component of the BRCA1/BRCA2 containing complex (BRCC), which also contains BRCA1, BRCA2, BARD1, BRCC3/BRCC36 and RAD51. BRCC is a ubiquitin E3 ligase complex that enhances cellular survival following DNA damage. May interact with FAS and TNFRSF1A.
Component of the ARISC complex, at least composed of UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:21282113, PubMed:24075985). Component of the BRCA1-A complex, at least composed of BRCA1, BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1. In the BRCA1-A complex, interacts directly with ABRAXAS1, BRCC3/BRCC36 and BABAM1/NBA1. Binds polyubiquitin. Component of the BRISC complex, at least composed of ABRAXAS2, BRCC3/BRCC36, BABAM2 and BABAM1/NBA1 (PubMed:19214193, PubMed:21282113, PubMed:24075985, PubMed:25283148). Identified in a complex with SHMT2 and the other subunits of the BRISC complex (PubMed:24075985). Component of the BRCA1/BRCA2 containing complex (BRCC), which also contains BRCA1, BRCA2, BARD1, BRCC3/BRCC36 and RAD51. BRCC is a ubiquitin E3 ligase complex that enhances cellular survival following DNA damage. May interact with FAS and TNFRSF1A (PubMed:15465831).
Similarity
Belongs to the BABAM2 family.
Keywords
Apoptosis
Cell cycle
Cell division
Chromatin regulator
Complete proteome
Cytoplasm
DNA damage
DNA repair
Mitosis
Nucleus
Reference proteome
Repeat
Ubl conjugation pathway
Acetylation
Phosphoprotein
Alternative splicing
Feature
chain BRISC and BRCA1-A complex member 2
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
A0A1E1WPJ3
A0A2H1VI71
A0A0L7KXZ0
A0A3S2LP93
A0A194R1C1
A0A212FFE3
+ More
A0A194PX89 S4PYE1 A0A2Z5TTJ8 A0A2J7QRH3 A0A067QST8 A0A210Q3P7 A0A1S3KD76 L7LSS0 A0A224YSG0 L7M1C1 A0A131YM22 A0A1E1XQJ9 A0A1E1XC56 V3ZRI7 E2AN93 G3MI65 A0A026VZI2 A0A131XEC3 A0A2P8XDL0 A0A195FT18 B6NXD5 F4X4N3 R7V964 K1PTQ4 A0A158NYZ8 A0A151WQR0 A0A131XUG7 K7F834 A0A087T7P8 W4YJ73 A0A091JIN3 Q6DJ78 A0A0L7RGQ9 A0A091I420 A0A091HFL5 A0A093CWL1 A0A091W1Z8 A0A091VUM6 A0A093GTB8 A0A093R1N9 A0A093JTT1 A0A087RAM7 A0A093J293 G1NDH0 A0A0K8RDG1 A0A093QJB4 A0A218UVW2 H0YWE9 G3URP2 A0A2I0URZ6 A0A2I0MAL9 Q5ZML0 A0A0A0A8A8 A0A091GX29 A0A384AVJ8 A0A1U7Q280 Q6SP92 A0A293LQR3 A0A091FVM5 U3K8E0 A0A1L8G6C3 A0A0Q3QYL6 A0A2K5SA17 E2RMQ2 A0A3Q7UWE0 A0A2K5SA41 A0A2J7QRF3 A0A2I2Z6N2 Q9NXR7-4 A0A2I3HWS4 A0A2R9BRV7 F7BJZ6 A0A2K6UJJ8 K7AYB2 Q5REX9 A0A2Y9S811 A0A2I2YFB1 A0A2I3GAP1 A0A2R9C0X7 H2P6L8 A0A2K6UJN9 F6YLX2 K7ALU2 Q8WN70 Q9NXR7 Q9NXR7-3 A0A1V4JJP4 A0A2K5SA35 A0A384CUH1 E2RMP1 F6PUR1 A0A0B8RTW4 D6X0Y3
A0A194PX89 S4PYE1 A0A2Z5TTJ8 A0A2J7QRH3 A0A067QST8 A0A210Q3P7 A0A1S3KD76 L7LSS0 A0A224YSG0 L7M1C1 A0A131YM22 A0A1E1XQJ9 A0A1E1XC56 V3ZRI7 E2AN93 G3MI65 A0A026VZI2 A0A131XEC3 A0A2P8XDL0 A0A195FT18 B6NXD5 F4X4N3 R7V964 K1PTQ4 A0A158NYZ8 A0A151WQR0 A0A131XUG7 K7F834 A0A087T7P8 W4YJ73 A0A091JIN3 Q6DJ78 A0A0L7RGQ9 A0A091I420 A0A091HFL5 A0A093CWL1 A0A091W1Z8 A0A091VUM6 A0A093GTB8 A0A093R1N9 A0A093JTT1 A0A087RAM7 A0A093J293 G1NDH0 A0A0K8RDG1 A0A093QJB4 A0A218UVW2 H0YWE9 G3URP2 A0A2I0URZ6 A0A2I0MAL9 Q5ZML0 A0A0A0A8A8 A0A091GX29 A0A384AVJ8 A0A1U7Q280 Q6SP92 A0A293LQR3 A0A091FVM5 U3K8E0 A0A1L8G6C3 A0A0Q3QYL6 A0A2K5SA17 E2RMQ2 A0A3Q7UWE0 A0A2K5SA41 A0A2J7QRF3 A0A2I2Z6N2 Q9NXR7-4 A0A2I3HWS4 A0A2R9BRV7 F7BJZ6 A0A2K6UJJ8 K7AYB2 Q5REX9 A0A2Y9S811 A0A2I2YFB1 A0A2I3GAP1 A0A2R9C0X7 H2P6L8 A0A2K6UJN9 F6YLX2 K7ALU2 Q8WN70 Q9NXR7 Q9NXR7-3 A0A1V4JJP4 A0A2K5SA35 A0A384CUH1 E2RMP1 F6PUR1 A0A0B8RTW4 D6X0Y3
Pubmed
26227816
26354079
22118469
23622113
26760975
24845553
+ More
28812685 25576852 28797301 26830274 29209593 28503490 23254933 20798317 22216098 24508170 30249741 28049606 29403074 18563158 21719571 22992520 21347285 17381049 20838655 20360741 23371554 15642098 15127289 27762356 16341006 22398555 7826398 11676476 14636569 14702039 15815621 15489334 15465831 17525341 19413330 19214193 19261746 19261749 19261748 20068231 21269460 21282113 22223895 22814378 24075985 23186163 25283148 26195665 22722832 16136131 25243066 24813606 18464734 18362917 19820115
28812685 25576852 28797301 26830274 29209593 28503490 23254933 20798317 22216098 24508170 30249741 28049606 29403074 18563158 21719571 22992520 21347285 17381049 20838655 20360741 23371554 15642098 15127289 27762356 16341006 22398555 7826398 11676476 14636569 14702039 15815621 15489334 15465831 17525341 19413330 19214193 19261746 19261749 19261748 20068231 21269460 21282113 22223895 22814378 24075985 23186163 25283148 26195665 22722832 16136131 25243066 24813606 18464734 18362917 19820115
EMBL
GDQN01002227
JAT88827.1
ODYU01002682
SOQ40491.1
JTDY01004644
KOB67916.1
+ More
RSAL01000040 RVE50957.1 KQ460883 KPJ11314.1 AGBW02008829 OWR52447.1 KQ459586 KPI97966.1 GAIX01003448 JAA89112.1 FX985788 BBA93675.1 NEVH01011897 PNF31170.1 KK853582 KDR06501.1 NEDP02005114 OWF43366.1 GACK01009918 JAA55116.1 GFPF01006385 MAA17531.1 GACK01006938 JAA58096.1 GEDV01008932 JAP79625.1 GFAA01002213 JAU01222.1 GFAC01002399 JAT96789.1 KB203660 ESO83491.1 GL441149 EFN65095.1 JO841566 AEO33183.1 KK107652 QOIP01000003 EZA48279.1 RLU24605.1 GEFH01002988 JAP65593.1 PYGN01002696 PSN30085.1 KQ981276 KYN43586.1 ABEP02001063 GL888668 EGI58587.1 AMQN01000776 KB295623 ELU12901.1 JH816684 EKC27637.1 ADTU01004416 KQ982821 KYQ50194.1 GEFM01004842 JAP70954.1 AGCU01036885 AGCU01036886 AGCU01036887 AGCU01036888 AGCU01036889 AGCU01036890 AGCU01036891 AGCU01036892 AGCU01036893 AGCU01036894 KK113841 KFM61137.1 AAGJ04016893 KK500912 KFP11536.1 BC075305 KQ414596 KOC70040.1 KL218170 KFP02932.1 KL536765 KFO94676.1 KL471104 KFV18873.1 KL411537 KFR08788.1 KK734339 KFR06465.1 KL216591 KFV70282.1 KL224848 KFW64939.1 KL206405 KFV81989.1 KL226263 KFM10531.1 KK573690 KFW09095.1 GADI01004616 JAA69192.1 KL761277 KFW89023.1 MUZQ01000117 OWK57766.1 ABQF01049147 ABQF01049148 ABQF01049149 ABQF01049150 KZ505646 PKU48825.1 AKCR02000024 PKK26730.1 AJ719374 KL870842 KGL89713.1 KL447916 KFO78657.1 AY454160 GFWV01005428 MAA30158.1 KK719710 KFO65215.1 AGTO01020624 CM004474 OCT79529.1 LMAW01002680 KQK78217.1 AAEX03010805 PNF31169.1 CABD030013680 CABD030013681 CABD030013682 CABD030013683 CABD030013684 CABD030013685 CABD030013686 CABD030013687 CABD030013688 CABD030013689 L38616 AF420605 AY438031 AF015767 AF420602 AF420603 AK000097 AK291086 AC021171 AC093690 AC096552 CH471053 BC001251 AAA64231.1 AAB69387.1 AAH01251.1 AAL17818.1 AAR30499.1 BAA90943.1 ADFV01011340 ADFV01011341 ADFV01011342 ADFV01011343 ADFV01011344 ADFV01011345 ADFV01011346 ADFV01011347 ADFV01011348 ADFV01011349 AJFE02095233 AJFE02095234 AJFE02095235 AJFE02095236 AJFE02095237 AJFE02095238 AJFE02095239 AJFE02095240 AJFE02095241 AJFE02095242 AACZ04002049 GABC01008590 GABF01001101 GABD01010119 GABE01004557 JAA02748.1 JAA21044.1 JAA22981.1 JAA40182.1 CR857384 ABGA01135681 ABGA01135682 ABGA01135683 ABGA01135684 ABGA01135685 ABGA01135686 ABGA01135687 ABGA01135688 ABGA01135689 ABGA01135690 GAMT01005345 GAMS01009383 GAMR01006517 GAMQ01007266 GAMP01002110 JAB06516.1 JAB13753.1 JAB27415.1 JAB34585.1 JAB50645.1 GABC01008592 GABC01008589 GABF01001103 GABD01010121 GABE01004559 JAA02749.1 JAA21042.1 JAA22979.1 JAA40180.1 AF440753 LSYS01007194 OPJ72399.1 GBZA01000078 JAG69690.1 KQ971372 EFA10575.1
RSAL01000040 RVE50957.1 KQ460883 KPJ11314.1 AGBW02008829 OWR52447.1 KQ459586 KPI97966.1 GAIX01003448 JAA89112.1 FX985788 BBA93675.1 NEVH01011897 PNF31170.1 KK853582 KDR06501.1 NEDP02005114 OWF43366.1 GACK01009918 JAA55116.1 GFPF01006385 MAA17531.1 GACK01006938 JAA58096.1 GEDV01008932 JAP79625.1 GFAA01002213 JAU01222.1 GFAC01002399 JAT96789.1 KB203660 ESO83491.1 GL441149 EFN65095.1 JO841566 AEO33183.1 KK107652 QOIP01000003 EZA48279.1 RLU24605.1 GEFH01002988 JAP65593.1 PYGN01002696 PSN30085.1 KQ981276 KYN43586.1 ABEP02001063 GL888668 EGI58587.1 AMQN01000776 KB295623 ELU12901.1 JH816684 EKC27637.1 ADTU01004416 KQ982821 KYQ50194.1 GEFM01004842 JAP70954.1 AGCU01036885 AGCU01036886 AGCU01036887 AGCU01036888 AGCU01036889 AGCU01036890 AGCU01036891 AGCU01036892 AGCU01036893 AGCU01036894 KK113841 KFM61137.1 AAGJ04016893 KK500912 KFP11536.1 BC075305 KQ414596 KOC70040.1 KL218170 KFP02932.1 KL536765 KFO94676.1 KL471104 KFV18873.1 KL411537 KFR08788.1 KK734339 KFR06465.1 KL216591 KFV70282.1 KL224848 KFW64939.1 KL206405 KFV81989.1 KL226263 KFM10531.1 KK573690 KFW09095.1 GADI01004616 JAA69192.1 KL761277 KFW89023.1 MUZQ01000117 OWK57766.1 ABQF01049147 ABQF01049148 ABQF01049149 ABQF01049150 KZ505646 PKU48825.1 AKCR02000024 PKK26730.1 AJ719374 KL870842 KGL89713.1 KL447916 KFO78657.1 AY454160 GFWV01005428 MAA30158.1 KK719710 KFO65215.1 AGTO01020624 CM004474 OCT79529.1 LMAW01002680 KQK78217.1 AAEX03010805 PNF31169.1 CABD030013680 CABD030013681 CABD030013682 CABD030013683 CABD030013684 CABD030013685 CABD030013686 CABD030013687 CABD030013688 CABD030013689 L38616 AF420605 AY438031 AF015767 AF420602 AF420603 AK000097 AK291086 AC021171 AC093690 AC096552 CH471053 BC001251 AAA64231.1 AAB69387.1 AAH01251.1 AAL17818.1 AAR30499.1 BAA90943.1 ADFV01011340 ADFV01011341 ADFV01011342 ADFV01011343 ADFV01011344 ADFV01011345 ADFV01011346 ADFV01011347 ADFV01011348 ADFV01011349 AJFE02095233 AJFE02095234 AJFE02095235 AJFE02095236 AJFE02095237 AJFE02095238 AJFE02095239 AJFE02095240 AJFE02095241 AJFE02095242 AACZ04002049 GABC01008590 GABF01001101 GABD01010119 GABE01004557 JAA02748.1 JAA21044.1 JAA22981.1 JAA40182.1 CR857384 ABGA01135681 ABGA01135682 ABGA01135683 ABGA01135684 ABGA01135685 ABGA01135686 ABGA01135687 ABGA01135688 ABGA01135689 ABGA01135690 GAMT01005345 GAMS01009383 GAMR01006517 GAMQ01007266 GAMP01002110 JAB06516.1 JAB13753.1 JAB27415.1 JAB34585.1 JAB50645.1 GABC01008592 GABC01008589 GABF01001103 GABD01010121 GABE01004559 JAA02749.1 JAA21042.1 JAA22979.1 JAA40180.1 AF440753 LSYS01007194 OPJ72399.1 GBZA01000078 JAG69690.1 KQ971372 EFA10575.1
Proteomes
UP000037510
UP000283053
UP000053240
UP000007151
UP000053268
UP000235965
+ More
UP000027135 UP000242188 UP000085678 UP000030746 UP000000311 UP000053097 UP000279307 UP000245037 UP000078541 UP000001554 UP000007755 UP000014760 UP000005408 UP000005205 UP000075809 UP000007267 UP000054359 UP000007110 UP000053119 UP000008143 UP000053825 UP000054308 UP000053283 UP000053605 UP000053875 UP000054081 UP000053584 UP000053286 UP000001645 UP000053258 UP000197619 UP000007754 UP000053872 UP000000539 UP000053858 UP000053760 UP000261681 UP000189706 UP000052976 UP000016665 UP000186698 UP000051836 UP000233040 UP000002254 UP000286640 UP000001519 UP000005640 UP000001073 UP000240080 UP000008225 UP000233220 UP000002277 UP000001595 UP000190648 UP000261680 UP000291021 UP000002279 UP000007266
UP000027135 UP000242188 UP000085678 UP000030746 UP000000311 UP000053097 UP000279307 UP000245037 UP000078541 UP000001554 UP000007755 UP000014760 UP000005408 UP000005205 UP000075809 UP000007267 UP000054359 UP000007110 UP000053119 UP000008143 UP000053825 UP000054308 UP000053283 UP000053605 UP000053875 UP000054081 UP000053584 UP000053286 UP000001645 UP000053258 UP000197619 UP000007754 UP000053872 UP000000539 UP000053858 UP000053760 UP000261681 UP000189706 UP000052976 UP000016665 UP000186698 UP000051836 UP000233040 UP000002254 UP000286640 UP000001519 UP000005640 UP000001073 UP000240080 UP000008225 UP000233220 UP000002277 UP000001595 UP000190648 UP000261680 UP000291021 UP000002279 UP000007266
Pfam
PF06113 BRE
SUPFAM
SSF54495
SSF54495
ProteinModelPortal
A0A1E1WPJ3
A0A2H1VI71
A0A0L7KXZ0
A0A3S2LP93
A0A194R1C1
A0A212FFE3
+ More
A0A194PX89 S4PYE1 A0A2Z5TTJ8 A0A2J7QRH3 A0A067QST8 A0A210Q3P7 A0A1S3KD76 L7LSS0 A0A224YSG0 L7M1C1 A0A131YM22 A0A1E1XQJ9 A0A1E1XC56 V3ZRI7 E2AN93 G3MI65 A0A026VZI2 A0A131XEC3 A0A2P8XDL0 A0A195FT18 B6NXD5 F4X4N3 R7V964 K1PTQ4 A0A158NYZ8 A0A151WQR0 A0A131XUG7 K7F834 A0A087T7P8 W4YJ73 A0A091JIN3 Q6DJ78 A0A0L7RGQ9 A0A091I420 A0A091HFL5 A0A093CWL1 A0A091W1Z8 A0A091VUM6 A0A093GTB8 A0A093R1N9 A0A093JTT1 A0A087RAM7 A0A093J293 G1NDH0 A0A0K8RDG1 A0A093QJB4 A0A218UVW2 H0YWE9 G3URP2 A0A2I0URZ6 A0A2I0MAL9 Q5ZML0 A0A0A0A8A8 A0A091GX29 A0A384AVJ8 A0A1U7Q280 Q6SP92 A0A293LQR3 A0A091FVM5 U3K8E0 A0A1L8G6C3 A0A0Q3QYL6 A0A2K5SA17 E2RMQ2 A0A3Q7UWE0 A0A2K5SA41 A0A2J7QRF3 A0A2I2Z6N2 Q9NXR7-4 A0A2I3HWS4 A0A2R9BRV7 F7BJZ6 A0A2K6UJJ8 K7AYB2 Q5REX9 A0A2Y9S811 A0A2I2YFB1 A0A2I3GAP1 A0A2R9C0X7 H2P6L8 A0A2K6UJN9 F6YLX2 K7ALU2 Q8WN70 Q9NXR7 Q9NXR7-3 A0A1V4JJP4 A0A2K5SA35 A0A384CUH1 E2RMP1 F6PUR1 A0A0B8RTW4 D6X0Y3
A0A194PX89 S4PYE1 A0A2Z5TTJ8 A0A2J7QRH3 A0A067QST8 A0A210Q3P7 A0A1S3KD76 L7LSS0 A0A224YSG0 L7M1C1 A0A131YM22 A0A1E1XQJ9 A0A1E1XC56 V3ZRI7 E2AN93 G3MI65 A0A026VZI2 A0A131XEC3 A0A2P8XDL0 A0A195FT18 B6NXD5 F4X4N3 R7V964 K1PTQ4 A0A158NYZ8 A0A151WQR0 A0A131XUG7 K7F834 A0A087T7P8 W4YJ73 A0A091JIN3 Q6DJ78 A0A0L7RGQ9 A0A091I420 A0A091HFL5 A0A093CWL1 A0A091W1Z8 A0A091VUM6 A0A093GTB8 A0A093R1N9 A0A093JTT1 A0A087RAM7 A0A093J293 G1NDH0 A0A0K8RDG1 A0A093QJB4 A0A218UVW2 H0YWE9 G3URP2 A0A2I0URZ6 A0A2I0MAL9 Q5ZML0 A0A0A0A8A8 A0A091GX29 A0A384AVJ8 A0A1U7Q280 Q6SP92 A0A293LQR3 A0A091FVM5 U3K8E0 A0A1L8G6C3 A0A0Q3QYL6 A0A2K5SA17 E2RMQ2 A0A3Q7UWE0 A0A2K5SA41 A0A2J7QRF3 A0A2I2Z6N2 Q9NXR7-4 A0A2I3HWS4 A0A2R9BRV7 F7BJZ6 A0A2K6UJJ8 K7AYB2 Q5REX9 A0A2Y9S811 A0A2I2YFB1 A0A2I3GAP1 A0A2R9C0X7 H2P6L8 A0A2K6UJN9 F6YLX2 K7ALU2 Q8WN70 Q9NXR7 Q9NXR7-3 A0A1V4JJP4 A0A2K5SA35 A0A384CUH1 E2RMP1 F6PUR1 A0A0B8RTW4 D6X0Y3
PDB
6R8F
E-value=7.72119e-11,
Score=161
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Localizes at sites of DNA damage at double-strand breaks (DSBs). With evidence from 1 publications.
Nucleus Localizes at sites of DNA damage at double-strand breaks (DSBs). With evidence from 1 publications.
Nucleus Localizes at sites of DNA damage at double-strand breaks (DSBs). With evidence from 1 publications.
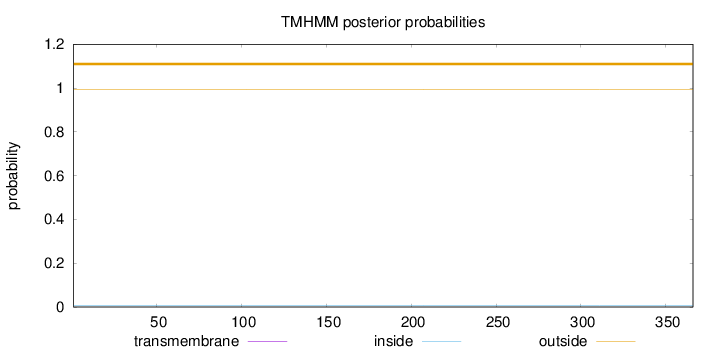
Length:
366
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000930000000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00526
outside
1 - 366
Population Genetic Test Statistics
Pi
250.927422
Theta
168.641013
Tajima's D
1.202973
CLR
0.502026
CSRT
0.709814509274536
Interpretation
Uncertain
