Pre Gene Modal
BGIBMGA009986
Annotation
PREDICTED:_uncharacterized_protein_YJR142W_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.453
Sequence
CDS
ATGAAAGATATGAATTCAGCACAAAACAACGTTTCGGATTTGTTAAAACTAGCCCGCAAGTTTAATTCTTTCTACCTCTCAGGTTTACATCAAGGAATATGTAAGCCCTTCATAGTGGCTGGGCATCAAGTTGGTCTTGTCCGACCTGATGTTTTAAAATATTTGCAGCGTTTCCCAGAGGTGTTCAGAATCGCCGGGAAATATGTGGAATTAAATCCTGCTTTTAGAGATTACAAAGAAAGGACCACAAGAGTTGCTGATGTCCTGCAAAAACTGAGGAAGGAGAATGAGATATGCGCATTAAAAGGATGGAGAGATGAGTGTTTTGAAGTCAGCACTGCGTTTTATCAAGAGAGCCTATTGGAGATGGACAGGAGTGCCATATGCCTATTTGGTATTAGAAATTATGGCGTCAGTGTCACCGGCTATCTCAATCATCCCAGTAAAGGACTATGCATTTGGTTACAGCAGCGGAGTTTTACCAAACAAACATGGCCTGGTAAATGGGATTGCTTTGTTAGCGGTGGTCTCGCAGTGGGATTTGGTATTCTAGAGACAACTATAAAAGAGGCCGCAGAAGAGGCATCTGTGGAGGGAGAATTGGTAAAAAAATTGGTACCAGCTGGCTGTGTAAGTTTCTATTTTGAAAGCGAGCGAGGCCTGTTTCCAAACACGGAGTACGTTTATGACCTTGAACTGCCTTTGGATTTCGTACCGCAAAACGCTGATGGCGAAGTCGAGAGCTTCGAGCTGCTCACTGCTGAGGAATGTATACAGCGAGCTCTCTCGCCTCAGTTCAAGACCACCAGTGCACCAGTACTCATAGATTTCCTCATACGTCGAGGTTATATCAATCCGGAGAACGAGCCAAACTATCGGTACTTGGTCGAGTTGTTGCACGTACCGTTGCAGTCTATTTACAATTGGCCGTTCACTCACATGGTCGCCGCGGCCGCCGTCAACGGAGACGACCAAGGCATATACGACGACAACTAA
Protein
MKDMNSAQNNVSDLLKLARKFNSFYLSGLHQGICKPFIVAGHQVGLVRPDVLKYLQRFPEVFRIAGKYVELNPAFRDYKERTTRVADVLQKLRKENEICALKGWRDECFEVSTAFYQESLLEMDRSAICLFGIRNYGVSVTGYLNHPSKGLCIWLQQRSFTKQTWPGKWDCFVSGGLAVGFGILETTIKEAAEEASVEGELVKKLVPAGCVSFYFESERGLFPNTEYVYDLELPLDFVPQNADGEVESFELLTAEECIQRALSPQFKTTSAPVLIDFLIRRGYINPENEPNYRYLVELLHVPLQSIYNWPFTHMVAAAAVNGDDQGIYDDN
Summary
Uniprot
H9JKD5
A0A2H1VI35
A0A1E1WAA4
A0A194Q3H6
A0A212FFE2
A0A194R236
+ More
A0A0L7KXB9 S4PRG7 A0A1Y1MYD6 A0A139WBT1 D6WZN1 A0A182JWN6 A0A182LT52 A0A084WHA8 A0A182W7H8 A0A182NGI6 A0A182QMW5 A0A182XZU7 A0A182PD04 A0A182J8W3 A0A182L1Z4 A0A1S4GXJ4 Q7Q3P1 A0A182WT45 A0A182VH86 A0A182HV40 A0A0K8TT81 A0A067RKF1 B4KFF7 A0A1L8DYB4 A0A2M4BTD4 A0A2M4APQ8 B4LQP9 A0A0K8UHR9 A0A1Q3F5J2 E2BPV7 W5JHW3 A0A182F0S5 A0A1Q3F5C1 A0A2M3ZAE3 A0A0M8ZNX8 A0A088ABQ5 A0A034WBW4 A0A2A3E5J8 B0WE28 F4WJN3 A0A1L8DXP8 A0A3L8DH24 A0A151WH13 A0A195DEB6 A0A0L7RK39 A0A195AUT4 A0A158NNC3 A0A0A1XKY2 A0A0J7L241 A0A195BZV1 E0VEI3 A0A182GMP9 J3JXW9 A0A026X421 Q7PLS1 Q16KA3 A0A3B0KBK9 A0A1B6GC62 A0A0J9R5W9 A0A1A9W5I8 B5DK41 T1PG60 A0A0Q5VXG5 B4IVB3 A0A1J1IX48 A0A2S2N902 A0A0M4EEG0 A0A0C9QEL6 J9JYV2 A0A2S2Q6J4 A0A0R3NV11 A0A0L0C322 A0A182RJA8 A0A232F1F4 A0A0P9AAL3 A0A182TGY5 E9IR35 A0A1I8QET2 W8AZ05 A0A0A9WYM3 A0A2P8ZDY2 B4MU73 A0A2M4APV4 U4UIR7 A0A182SJX4 A0A2M4BU85 A0A0R1E8J3 A0A1A9VF35 E2AWE6 A0A1A9XQQ6 A0A1B0BN38 B4JQ25 A0A1A9ZFB8
A0A0L7KXB9 S4PRG7 A0A1Y1MYD6 A0A139WBT1 D6WZN1 A0A182JWN6 A0A182LT52 A0A084WHA8 A0A182W7H8 A0A182NGI6 A0A182QMW5 A0A182XZU7 A0A182PD04 A0A182J8W3 A0A182L1Z4 A0A1S4GXJ4 Q7Q3P1 A0A182WT45 A0A182VH86 A0A182HV40 A0A0K8TT81 A0A067RKF1 B4KFF7 A0A1L8DYB4 A0A2M4BTD4 A0A2M4APQ8 B4LQP9 A0A0K8UHR9 A0A1Q3F5J2 E2BPV7 W5JHW3 A0A182F0S5 A0A1Q3F5C1 A0A2M3ZAE3 A0A0M8ZNX8 A0A088ABQ5 A0A034WBW4 A0A2A3E5J8 B0WE28 F4WJN3 A0A1L8DXP8 A0A3L8DH24 A0A151WH13 A0A195DEB6 A0A0L7RK39 A0A195AUT4 A0A158NNC3 A0A0A1XKY2 A0A0J7L241 A0A195BZV1 E0VEI3 A0A182GMP9 J3JXW9 A0A026X421 Q7PLS1 Q16KA3 A0A3B0KBK9 A0A1B6GC62 A0A0J9R5W9 A0A1A9W5I8 B5DK41 T1PG60 A0A0Q5VXG5 B4IVB3 A0A1J1IX48 A0A2S2N902 A0A0M4EEG0 A0A0C9QEL6 J9JYV2 A0A2S2Q6J4 A0A0R3NV11 A0A0L0C322 A0A182RJA8 A0A232F1F4 A0A0P9AAL3 A0A182TGY5 E9IR35 A0A1I8QET2 W8AZ05 A0A0A9WYM3 A0A2P8ZDY2 B4MU73 A0A2M4APV4 U4UIR7 A0A182SJX4 A0A2M4BU85 A0A0R1E8J3 A0A1A9VF35 E2AWE6 A0A1A9XQQ6 A0A1B0BN38 B4JQ25 A0A1A9ZFB8
Pubmed
19121390
26354079
22118469
26227816
23622113
28004739
+ More
18362917 19820115 24438588 25244985 20966253 12364791 26369729 24845553 17994087 20798317 20920257 23761445 25348373 21719571 30249741 21347285 25830018 20566863 26483478 22516182 23537049 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 22936249 15632085 25315136 17550304 26108605 28648823 21282665 24495485 25401762 29403074
18362917 19820115 24438588 25244985 20966253 12364791 26369729 24845553 17994087 20798317 20920257 23761445 25348373 21719571 30249741 21347285 25830018 20566863 26483478 22516182 23537049 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 22936249 15632085 25315136 17550304 26108605 28648823 21282665 24495485 25401762 29403074
EMBL
BABH01036770
BABH01036771
ODYU01002682
SOQ40490.1
GDQN01007150
JAT83904.1
+ More
KQ459586 KPI97965.1 AGBW02008829 OWR52448.1 KQ460883 KPJ11315.1 JTDY01004644 KOB67918.1 GAIX01000810 JAA91750.1 GEZM01020098 JAV89405.1 KQ971372 KYB25360.1 EFA10452.2 AXCM01002571 ATLV01023800 KE525346 KFB49602.1 AXCN02000795 AAAB01008964 EAA12844.4 APCN01002493 GDAI01000247 JAI17356.1 KK852572 KDR21080.1 CH933807 EDW12057.1 GFDF01002839 JAV11245.1 GGFJ01006887 MBW56028.1 GGFK01009267 MBW42588.1 CH940649 EDW63433.1 GDHF01026196 JAI26118.1 GFDL01012272 JAV22773.1 GL449658 EFN82308.1 ADMH02001143 ETN63952.1 GFDL01012285 JAV22760.1 GGFM01004721 MBW25472.1 KQ435949 KOX68130.1 GAKP01007150 JAC51802.1 KZ288361 PBC26968.1 DS231904 EDS45204.1 GL888186 EGI65525.1 GFDF01002847 JAV11237.1 QOIP01000008 RLU19727.1 KQ983136 KYQ47139.1 KQ980989 KYN10769.1 KQ414575 KOC71235.1 KQ976738 KYM75822.1 ADTU01021247 GBXI01015996 GBXI01002641 JAC98295.1 JAD11651.1 LBMM01001129 KMQ96867.1 KQ978457 KYM93870.1 DS235092 EEB11789.1 JXUM01074933 JXUM01074934 JXUM01074935 KQ562859 KXJ74985.1 APGK01050705 BT128091 KB741175 AEE63052.1 ENN73216.1 KK107031 EZA62104.1 BT083411 AE014134 ACQ89820.1 EAA46065.2 EYR77321.1 CH477970 EAT34728.1 OUUW01000010 SPP85550.1 GECZ01009741 JAS60028.1 CM002910 KMY91498.1 CH379061 EDY70667.2 KA647746 AFP62375.1 CH954179 KQS61762.1 CH892674 EDX00278.2 CVRI01000061 CRL04132.1 GGMR01000793 MBY13412.1 CP012523 ALC40410.1 GBYB01001719 JAG71486.1 ABLF02039123 GGMS01004146 MBY73349.1 KRT04952.1 JRES01000973 KNC26642.1 NNAY01001273 OXU24544.1 CH902628 KPU75321.1 GL765043 EFZ16981.1 GAMC01015108 JAB91447.1 GBHO01033654 GBRD01015166 JAG09950.1 JAG50660.1 PYGN01000085 PSN54722.1 CH963852 EDW75662.1 GGFK01009317 MBW42638.1 KB632191 ERL89815.1 GGFJ01007433 MBW56574.1 KRK05533.1 GL443286 EFN62246.1 JXJN01017195 CH916372 EDV99005.1
KQ459586 KPI97965.1 AGBW02008829 OWR52448.1 KQ460883 KPJ11315.1 JTDY01004644 KOB67918.1 GAIX01000810 JAA91750.1 GEZM01020098 JAV89405.1 KQ971372 KYB25360.1 EFA10452.2 AXCM01002571 ATLV01023800 KE525346 KFB49602.1 AXCN02000795 AAAB01008964 EAA12844.4 APCN01002493 GDAI01000247 JAI17356.1 KK852572 KDR21080.1 CH933807 EDW12057.1 GFDF01002839 JAV11245.1 GGFJ01006887 MBW56028.1 GGFK01009267 MBW42588.1 CH940649 EDW63433.1 GDHF01026196 JAI26118.1 GFDL01012272 JAV22773.1 GL449658 EFN82308.1 ADMH02001143 ETN63952.1 GFDL01012285 JAV22760.1 GGFM01004721 MBW25472.1 KQ435949 KOX68130.1 GAKP01007150 JAC51802.1 KZ288361 PBC26968.1 DS231904 EDS45204.1 GL888186 EGI65525.1 GFDF01002847 JAV11237.1 QOIP01000008 RLU19727.1 KQ983136 KYQ47139.1 KQ980989 KYN10769.1 KQ414575 KOC71235.1 KQ976738 KYM75822.1 ADTU01021247 GBXI01015996 GBXI01002641 JAC98295.1 JAD11651.1 LBMM01001129 KMQ96867.1 KQ978457 KYM93870.1 DS235092 EEB11789.1 JXUM01074933 JXUM01074934 JXUM01074935 KQ562859 KXJ74985.1 APGK01050705 BT128091 KB741175 AEE63052.1 ENN73216.1 KK107031 EZA62104.1 BT083411 AE014134 ACQ89820.1 EAA46065.2 EYR77321.1 CH477970 EAT34728.1 OUUW01000010 SPP85550.1 GECZ01009741 JAS60028.1 CM002910 KMY91498.1 CH379061 EDY70667.2 KA647746 AFP62375.1 CH954179 KQS61762.1 CH892674 EDX00278.2 CVRI01000061 CRL04132.1 GGMR01000793 MBY13412.1 CP012523 ALC40410.1 GBYB01001719 JAG71486.1 ABLF02039123 GGMS01004146 MBY73349.1 KRT04952.1 JRES01000973 KNC26642.1 NNAY01001273 OXU24544.1 CH902628 KPU75321.1 GL765043 EFZ16981.1 GAMC01015108 JAB91447.1 GBHO01033654 GBRD01015166 JAG09950.1 JAG50660.1 PYGN01000085 PSN54722.1 CH963852 EDW75662.1 GGFK01009317 MBW42638.1 KB632191 ERL89815.1 GGFJ01007433 MBW56574.1 KRK05533.1 GL443286 EFN62246.1 JXJN01017195 CH916372 EDV99005.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000037510
UP000007266
+ More
UP000075881 UP000075883 UP000030765 UP000075920 UP000075884 UP000075886 UP000076408 UP000075885 UP000075880 UP000075882 UP000007062 UP000076407 UP000075903 UP000075840 UP000027135 UP000009192 UP000008792 UP000008237 UP000000673 UP000069272 UP000053105 UP000005203 UP000242457 UP000002320 UP000007755 UP000279307 UP000075809 UP000078492 UP000053825 UP000078540 UP000005205 UP000036403 UP000078542 UP000009046 UP000069940 UP000249989 UP000019118 UP000053097 UP000000803 UP000008820 UP000268350 UP000091820 UP000001819 UP000095301 UP000008711 UP000002282 UP000183832 UP000092553 UP000007819 UP000037069 UP000075900 UP000215335 UP000007801 UP000075902 UP000095300 UP000245037 UP000007798 UP000030742 UP000075901 UP000078200 UP000000311 UP000092443 UP000092460 UP000001070 UP000092445
UP000075881 UP000075883 UP000030765 UP000075920 UP000075884 UP000075886 UP000076408 UP000075885 UP000075880 UP000075882 UP000007062 UP000076407 UP000075903 UP000075840 UP000027135 UP000009192 UP000008792 UP000008237 UP000000673 UP000069272 UP000053105 UP000005203 UP000242457 UP000002320 UP000007755 UP000279307 UP000075809 UP000078492 UP000053825 UP000078540 UP000005205 UP000036403 UP000078542 UP000009046 UP000069940 UP000249989 UP000019118 UP000053097 UP000000803 UP000008820 UP000268350 UP000091820 UP000001819 UP000095301 UP000008711 UP000002282 UP000183832 UP000092553 UP000007819 UP000037069 UP000075900 UP000215335 UP000007801 UP000075902 UP000095300 UP000245037 UP000007798 UP000030742 UP000075901 UP000078200 UP000000311 UP000092443 UP000092460 UP000001070 UP000092445
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JKD5
A0A2H1VI35
A0A1E1WAA4
A0A194Q3H6
A0A212FFE2
A0A194R236
+ More
A0A0L7KXB9 S4PRG7 A0A1Y1MYD6 A0A139WBT1 D6WZN1 A0A182JWN6 A0A182LT52 A0A084WHA8 A0A182W7H8 A0A182NGI6 A0A182QMW5 A0A182XZU7 A0A182PD04 A0A182J8W3 A0A182L1Z4 A0A1S4GXJ4 Q7Q3P1 A0A182WT45 A0A182VH86 A0A182HV40 A0A0K8TT81 A0A067RKF1 B4KFF7 A0A1L8DYB4 A0A2M4BTD4 A0A2M4APQ8 B4LQP9 A0A0K8UHR9 A0A1Q3F5J2 E2BPV7 W5JHW3 A0A182F0S5 A0A1Q3F5C1 A0A2M3ZAE3 A0A0M8ZNX8 A0A088ABQ5 A0A034WBW4 A0A2A3E5J8 B0WE28 F4WJN3 A0A1L8DXP8 A0A3L8DH24 A0A151WH13 A0A195DEB6 A0A0L7RK39 A0A195AUT4 A0A158NNC3 A0A0A1XKY2 A0A0J7L241 A0A195BZV1 E0VEI3 A0A182GMP9 J3JXW9 A0A026X421 Q7PLS1 Q16KA3 A0A3B0KBK9 A0A1B6GC62 A0A0J9R5W9 A0A1A9W5I8 B5DK41 T1PG60 A0A0Q5VXG5 B4IVB3 A0A1J1IX48 A0A2S2N902 A0A0M4EEG0 A0A0C9QEL6 J9JYV2 A0A2S2Q6J4 A0A0R3NV11 A0A0L0C322 A0A182RJA8 A0A232F1F4 A0A0P9AAL3 A0A182TGY5 E9IR35 A0A1I8QET2 W8AZ05 A0A0A9WYM3 A0A2P8ZDY2 B4MU73 A0A2M4APV4 U4UIR7 A0A182SJX4 A0A2M4BU85 A0A0R1E8J3 A0A1A9VF35 E2AWE6 A0A1A9XQQ6 A0A1B0BN38 B4JQ25 A0A1A9ZFB8
A0A0L7KXB9 S4PRG7 A0A1Y1MYD6 A0A139WBT1 D6WZN1 A0A182JWN6 A0A182LT52 A0A084WHA8 A0A182W7H8 A0A182NGI6 A0A182QMW5 A0A182XZU7 A0A182PD04 A0A182J8W3 A0A182L1Z4 A0A1S4GXJ4 Q7Q3P1 A0A182WT45 A0A182VH86 A0A182HV40 A0A0K8TT81 A0A067RKF1 B4KFF7 A0A1L8DYB4 A0A2M4BTD4 A0A2M4APQ8 B4LQP9 A0A0K8UHR9 A0A1Q3F5J2 E2BPV7 W5JHW3 A0A182F0S5 A0A1Q3F5C1 A0A2M3ZAE3 A0A0M8ZNX8 A0A088ABQ5 A0A034WBW4 A0A2A3E5J8 B0WE28 F4WJN3 A0A1L8DXP8 A0A3L8DH24 A0A151WH13 A0A195DEB6 A0A0L7RK39 A0A195AUT4 A0A158NNC3 A0A0A1XKY2 A0A0J7L241 A0A195BZV1 E0VEI3 A0A182GMP9 J3JXW9 A0A026X421 Q7PLS1 Q16KA3 A0A3B0KBK9 A0A1B6GC62 A0A0J9R5W9 A0A1A9W5I8 B5DK41 T1PG60 A0A0Q5VXG5 B4IVB3 A0A1J1IX48 A0A2S2N902 A0A0M4EEG0 A0A0C9QEL6 J9JYV2 A0A2S2Q6J4 A0A0R3NV11 A0A0L0C322 A0A182RJA8 A0A232F1F4 A0A0P9AAL3 A0A182TGY5 E9IR35 A0A1I8QET2 W8AZ05 A0A0A9WYM3 A0A2P8ZDY2 B4MU73 A0A2M4APV4 U4UIR7 A0A182SJX4 A0A2M4BU85 A0A0R1E8J3 A0A1A9VF35 E2AWE6 A0A1A9XQQ6 A0A1B0BN38 B4JQ25 A0A1A9ZFB8
PDB
3DUP
E-value=1.40431e-41,
Score=426
Ontologies
GO
Topology
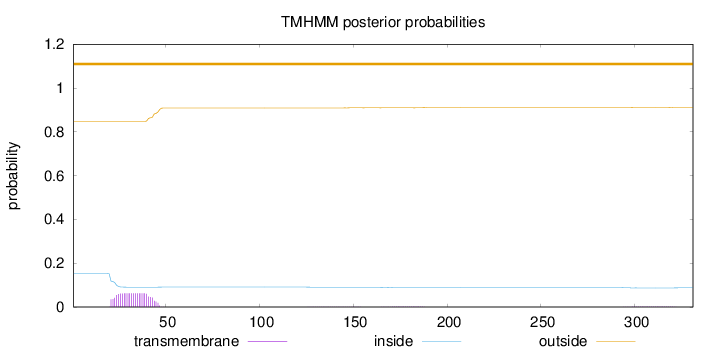
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.47086
Exp number, first 60 AAs:
1.37089
Total prob of N-in:
0.15251
outside
1 - 331
Population Genetic Test Statistics
Pi
260.763525
Theta
166.359987
Tajima's D
1.813412
CLR
0.424798
CSRT
0.850007499625019
Interpretation
Uncertain
