Gene
KWMTBOMO13215
Pre Gene Modal
BGIBMGA009962
Annotation
PREDICTED:_cAMP-specific_3'?5'-cyclic_phosphodiesterase?_isoforms_N/G_isoform_X3_[Bombyx_mori]
Full name
Phosphodiesterase
+ More
cAMP-specific 3',5'-cyclic phosphodiesterase
cAMP-specific 3',5'-cyclic phosphodiesterase, isoforms N/G
cAMP-specific 3',5'-cyclic phosphodiesterase, isoform I
cAMP-specific 3',5'-cyclic phosphodiesterase, isoform F
cAMP-specific 3',5'-cyclic phosphodiesterase
cAMP-specific 3',5'-cyclic phosphodiesterase, isoforms N/G
cAMP-specific 3',5'-cyclic phosphodiesterase, isoform I
cAMP-specific 3',5'-cyclic phosphodiesterase, isoform F
Alternative Name
Learning/memory process protein
Protein dunce
Protein dunce
Location in the cell
Nuclear Reliability : 2.599
Sequence
CDS
ATGAAGCTGGCTTTGGAAACCGTTGAGGAACTCGACTGGTGCCTGGACCAATTGGAGACGATACAGACACATCGATCGGTCTCCGACATGGCTTCGCTTAAGTTCAAGAGGATGCTGAACAAGGAGCTGAGCCACTTCTCAGAGTCCAGCAAGTCCGGCAATCAGATCTCCGAGTATATCTGCTCCACATTTTTGGACAAACAACAGGAGCTTGACTTACCATCCTTACAAGTCGAAGAAACTTCGGAACGCGGCGCCAAGAAAAAGGAAAAACCACGTCATGGCGGACCAGCGACTACGATGTCACAAATATCGGGTGTTAAGAGGACTTTGACACACACAAATAGCTTCACCGGCGAACGCGTGCCGAGGTATGGAGTTGAAACACCGCATGAGGAAGAGCTCGGCCTTCTTCTCAGCGATCTAGACCGGTGGGGAGTGAATATATTCCGCATCGGTGATCTGTCATGCGGTAGACCACTCACTGCTGTCGCGTACACAGCGTTCACATCACGGGAGCTGTTGACTACTCTTCAGATACCGGCGCGTACTTTCTTGGCGTTCGCAGTTACCTTAGAGGAGCACTATATTCGAGACAATCCTTTCCACAACTCTCTCCACGCTGCCGATGTCACACAGTCTACAAACGTACTGTTGAATACACCTGCGCTTAATGCTGTCTTCACACCACTAGAAGTTTGCGCTGCTCTTTTCGCTGCCTGCGTCCACGACGTCGATCACCCGGGCCTCACCAATCAATTCCTCGTCAACTCCAGTTCCGAACTCGCACTAATGTACAACGACGAGTCAGTCCTCGAGAACCACCACCTTGCAGTCGCATTCAAGCTGCTGCAGAATGATGGATGTGATATTTTCGTGAATCTTCACAAGAAACAAAGACAGACCCTCAGGAAAATGGTTATCGACATGGTCCTCTCGACAGATATGTCGAAACACATGAGTCTGCTGGCGGATCTGAAGACGATGGTCGAAACGAAGAAAGTCGCGGGTAGCGGTGTCTTATTGCTAGACAACTACACTGATAGAATTCAAGTATTAGAGAATCTCGTACACTGTGCCGATCTGAGCAATCCTACTAAGCCGTTGCCACTGTATAAACGATGGGTGGCATTACTGATGGAAGAGTTCTTCCAGCAAGGCGATCGTGAACGTGAACATGGCATGGATATCAGTCCAATGTGTGACAGACATAATGCTACTATAGAAAAATCTCAAGTCGGTTTCATTGACTATATCGTGCATCCTCTGTGGGAAACTTGGGCAGATTTGGTACATCCTGACGCTCAAGATATTTTGGACACCTTAGAAGAAAACCGAGACTATTACCAAAGTATGATTCCACCGTCACCGCCACCAGCACCGATACAATCCTCACACGCCTCTGATGATGATCGCCCTGAACAAATACGATTTCAAGTCACCCTAGAGGAGGGAGAAGGATCAGAAGATTGCGAGGGCGAAGGTGATGGTGGAATGTGA
Protein
MKLALETVEELDWCLDQLETIQTHRSVSDMASLKFKRMLNKELSHFSESSKSGNQISEYICSTFLDKQQELDLPSLQVEETSERGAKKKEKPRHGGPATTMSQISGVKRTLTHTNSFTGERVPRYGVETPHEEELGLLLSDLDRWGVNIFRIGDLSCGRPLTAVAYTAFTSRELLTTLQIPARTFLAFAVTLEEHYIRDNPFHNSLHAADVTQSTNVLLNTPALNAVFTPLEVCAALFAACVHDVDHPGLTNQFLVNSSSELALMYNDESVLENHHLAVAFKLLQNDGCDIFVNLHKKQRQTLRKMVIDMVLSTDMSKHMSLLADLKTMVETKKVAGSGVLLLDNYTDRIQVLENLVHCADLSNPTKPLPLYKRWVALLMEEFFQQGDREREHGMDISPMCDRHNATIEKSQVGFIDYIVHPLWETWADLVHPDAQDILDTLEENRDYYQSMIPPSPPPAPIQSSHASDDDRPEQIRFQVTLEEGEGSEDCEGEGDGGM
Summary
Description
Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes (By similarity). Vital for female fertility. Required for learning/memory.
Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes (By similarity). Vital for female fertility. Required for learning/memory (By similarity).
Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes (By similarity). Vital for female fertility. Required for learning/memory (By similarity).
Catalytic Activity
3',5'-cyclic AMP + H2O = AMP + H(+)
Cofactor
a divalent metal cation
Subunit
Monomer.
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Belongs to the cyclic nucleotide phosphodiesterase family. PDE4 subfamily.
Belongs to the cyclic nucleotide phosphodiesterase family. PDE4 subfamily.
Keywords
Alternative initiation
Alternative splicing
cAMP
Complete proteome
Hydrolase
Metal-binding
Reference proteome
Feature
chain Phosphodiesterase
splice variant In isoform VII.
splice variant In isoform VII.
Uniprot
A0A1E1W0K8
H9JKB1
A0A1E1W093
A0A194R246
A0A2H1W3R2
A0A194Q3G7
+ More
A0A212ENZ6 A0A1B0CBX1 A0A2A3E5J0 V9I967 B4JMU1 A0A3B0K0B8 V9IAZ4 A0A0Q9W6Z7 A0A0Q9WB84 A0A0M4F9F4 A0A0Q9VZ20 A0A1I8QDE8 A0A1I8QD96 A0A1I8QD92 A0A1L8DLH8 A0A1B6ERJ9 A0A1I8QDE4 A0A1I8QDA1 A0A1A9ZLK7 A0A1L8DLL1 A0A0Q9WZA2 A0A0Q9WNL3 A0A1B6FHL2 A0A1L8DL56 P12252-5 A0A1I8MIT5 E2QD73 A0A0Q9VZ02 A0A1I8QD76 P12252 P12252-4 A0A0Q9VZB9 A0A1I8QDA8 B4MJA8 E2AWF4 A0A1L8DLD7 A0A195FWK9 B4MCU3 A0A0Q9WBB8 A0A1A9WIK4 A0A1W4W882 A0A0Q9WNT8 A0A1W4WIJ3 A0A0Q5TGS1 A0A0Q5T381 A0A1W4VSN5 A0A0Q9VZ07 A0A1W4WJS5 A0A1W4VEP6 A0A1W4VEE2 A0A0Q9WAJ9 A0A1W4W883 A0A0T6AVH7 A0A0Q5TG56 Q9W4S9-2 A0A154P7D8 D0IQA0 K7J1X9 A0A1I8QD97 A0A1I8QDB2 B4IA32 A0A1B6E622 A0A0Q9WNR7 A0A151WH39 Q9W4S9 A0A1I8QD91 Q8MS17 T1PME5 H8F4P8 Q9W4T4 A0A1W4WJI5 A0A1W4WIJ8 A0A1B6DRH6 A0A1W4VRE9 A0A1W4VS49 A0A1W4WJJ1 A0A1W4W887 A0A1B6CIC6 A0A1B6CZC8 P12252-7 A0A232FBZ1 A0A1W4W888 M9PDN2 P12252-9 A0A1W4WJT5 C9QPC1 A0A0L0C0L2 Q8IRU4 A0A1W4W891 A0A0C9PRG4 B3N1U9 A0A0Q5T2H8 A0A195C111 P12252-8
A0A212ENZ6 A0A1B0CBX1 A0A2A3E5J0 V9I967 B4JMU1 A0A3B0K0B8 V9IAZ4 A0A0Q9W6Z7 A0A0Q9WB84 A0A0M4F9F4 A0A0Q9VZ20 A0A1I8QDE8 A0A1I8QD96 A0A1I8QD92 A0A1L8DLH8 A0A1B6ERJ9 A0A1I8QDE4 A0A1I8QDA1 A0A1A9ZLK7 A0A1L8DLL1 A0A0Q9WZA2 A0A0Q9WNL3 A0A1B6FHL2 A0A1L8DL56 P12252-5 A0A1I8MIT5 E2QD73 A0A0Q9VZ02 A0A1I8QD76 P12252 P12252-4 A0A0Q9VZB9 A0A1I8QDA8 B4MJA8 E2AWF4 A0A1L8DLD7 A0A195FWK9 B4MCU3 A0A0Q9WBB8 A0A1A9WIK4 A0A1W4W882 A0A0Q9WNT8 A0A1W4WIJ3 A0A0Q5TGS1 A0A0Q5T381 A0A1W4VSN5 A0A0Q9VZ07 A0A1W4WJS5 A0A1W4VEP6 A0A1W4VEE2 A0A0Q9WAJ9 A0A1W4W883 A0A0T6AVH7 A0A0Q5TG56 Q9W4S9-2 A0A154P7D8 D0IQA0 K7J1X9 A0A1I8QD97 A0A1I8QDB2 B4IA32 A0A1B6E622 A0A0Q9WNR7 A0A151WH39 Q9W4S9 A0A1I8QD91 Q8MS17 T1PME5 H8F4P8 Q9W4T4 A0A1W4WJI5 A0A1W4WIJ8 A0A1B6DRH6 A0A1W4VRE9 A0A1W4VS49 A0A1W4WJJ1 A0A1W4W887 A0A1B6CIC6 A0A1B6CZC8 P12252-7 A0A232FBZ1 A0A1W4W888 M9PDN2 P12252-9 A0A1W4WJT5 C9QPC1 A0A0L0C0L2 Q8IRU4 A0A1W4W891 A0A0C9PRG4 B3N1U9 A0A0Q5T2H8 A0A195C111 P12252-8
EC Number
3.1.4.-
3.1.4.53
3.1.4.53
Pubmed
EMBL
GDQN01010548
JAT80506.1
BABH01036773
GDQN01010644
JAT80410.1
KQ460883
+ More
KPJ11325.1 ODYU01005845 SOQ47134.1 KQ459586 KPI97955.1 AGBW02013576 OWR43204.1 AJWK01005928 AJWK01005929 AJWK01005930 AJWK01005931 KZ288361 PBC26965.1 JR037164 JR037167 AEY57620.1 AEY57622.1 CH916371 EDV92034.1 OUUW01000003 SPP78451.1 JR037165 JR037168 AEY57621.1 AEY57623.1 CH940660 KRF78071.1 KRF78075.1 CP012528 ALC48832.1 KRF78074.1 GFDF01006884 JAV07200.1 GECZ01029271 JAS40498.1 GFDF01006829 JAV07255.1 CH963738 KRF97512.1 KRF97513.1 GECZ01020080 GECZ01000476 JAS49689.1 JAS69293.1 GFDF01006885 JAV07199.1 X55167 X55168 X55169 X55170 X55171 X55172 X55173 X55174 X55175 M14982 M14978 M14979 M14980 M14981 AE014298 AL121800 AY119511 AAN09603.2 KRF78076.1 KRF78077.1 EDW72197.2 GL443286 EFN62254.1 GFDF01006899 JAV07185.1 KQ981208 KYN44697.1 EDW58015.1 KRF78078.1 KRF97511.1 CH954180 KQS29675.1 KQS29679.1 KRF78073.1 KRF78079.1 LJIG01022741 KRT78957.1 KQS29678.1 AAF45863.2 KQ434832 KZC07855.1 BT100138 ACX94163.1 CH480825 EDW44063.1 GEDC01003926 JAS33372.1 KRF97510.1 KQ983136 KYQ47128.1 AY119147 AAM51007.1 KA644518 KA649083 AFP63712.1 BT133288 AAN09599.1 AFC88877.1 AL024484 AAF45858.3 GEDC01023956 GEDC01009028 JAS13342.1 JAS28270.1 GEDC01024213 JAS13085.1 GEDC01018496 JAS18802.1 NNAY01000471 OXU28142.1 AGB95047.1 BT100003 ACX54887.1 JRES01001065 KNC25848.1 AAN09606.1 GBYB01003893 JAG73660.1 CH902663 EDV29990.1 KQS29673.1 KQ978457 KYM93858.1
KPJ11325.1 ODYU01005845 SOQ47134.1 KQ459586 KPI97955.1 AGBW02013576 OWR43204.1 AJWK01005928 AJWK01005929 AJWK01005930 AJWK01005931 KZ288361 PBC26965.1 JR037164 JR037167 AEY57620.1 AEY57622.1 CH916371 EDV92034.1 OUUW01000003 SPP78451.1 JR037165 JR037168 AEY57621.1 AEY57623.1 CH940660 KRF78071.1 KRF78075.1 CP012528 ALC48832.1 KRF78074.1 GFDF01006884 JAV07200.1 GECZ01029271 JAS40498.1 GFDF01006829 JAV07255.1 CH963738 KRF97512.1 KRF97513.1 GECZ01020080 GECZ01000476 JAS49689.1 JAS69293.1 GFDF01006885 JAV07199.1 X55167 X55168 X55169 X55170 X55171 X55172 X55173 X55174 X55175 M14982 M14978 M14979 M14980 M14981 AE014298 AL121800 AY119511 AAN09603.2 KRF78076.1 KRF78077.1 EDW72197.2 GL443286 EFN62254.1 GFDF01006899 JAV07185.1 KQ981208 KYN44697.1 EDW58015.1 KRF78078.1 KRF97511.1 CH954180 KQS29675.1 KQS29679.1 KRF78073.1 KRF78079.1 LJIG01022741 KRT78957.1 KQS29678.1 AAF45863.2 KQ434832 KZC07855.1 BT100138 ACX94163.1 CH480825 EDW44063.1 GEDC01003926 JAS33372.1 KRF97510.1 KQ983136 KYQ47128.1 AY119147 AAM51007.1 KA644518 KA649083 AFP63712.1 BT133288 AAN09599.1 AFC88877.1 AL024484 AAF45858.3 GEDC01023956 GEDC01009028 JAS13342.1 JAS28270.1 GEDC01024213 JAS13085.1 GEDC01018496 JAS18802.1 NNAY01000471 OXU28142.1 AGB95047.1 BT100003 ACX54887.1 JRES01001065 KNC25848.1 AAN09606.1 GBYB01003893 JAG73660.1 CH902663 EDV29990.1 KQS29673.1 KQ978457 KYM93858.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000092461
UP000242457
+ More
UP000001070 UP000268350 UP000008792 UP000092553 UP000095300 UP000092445 UP000007798 UP000000803 UP000095301 UP000000311 UP000078541 UP000091820 UP000192223 UP000008711 UP000192221 UP000076502 UP000002358 UP000001292 UP000075809 UP000215335 UP000037069 UP000007801 UP000078542
UP000001070 UP000268350 UP000008792 UP000092553 UP000095300 UP000092445 UP000007798 UP000000803 UP000095301 UP000000311 UP000078541 UP000091820 UP000192223 UP000008711 UP000192221 UP000076502 UP000002358 UP000001292 UP000075809 UP000215335 UP000037069 UP000007801 UP000078542
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A1E1W0K8
H9JKB1
A0A1E1W093
A0A194R246
A0A2H1W3R2
A0A194Q3G7
+ More
A0A212ENZ6 A0A1B0CBX1 A0A2A3E5J0 V9I967 B4JMU1 A0A3B0K0B8 V9IAZ4 A0A0Q9W6Z7 A0A0Q9WB84 A0A0M4F9F4 A0A0Q9VZ20 A0A1I8QDE8 A0A1I8QD96 A0A1I8QD92 A0A1L8DLH8 A0A1B6ERJ9 A0A1I8QDE4 A0A1I8QDA1 A0A1A9ZLK7 A0A1L8DLL1 A0A0Q9WZA2 A0A0Q9WNL3 A0A1B6FHL2 A0A1L8DL56 P12252-5 A0A1I8MIT5 E2QD73 A0A0Q9VZ02 A0A1I8QD76 P12252 P12252-4 A0A0Q9VZB9 A0A1I8QDA8 B4MJA8 E2AWF4 A0A1L8DLD7 A0A195FWK9 B4MCU3 A0A0Q9WBB8 A0A1A9WIK4 A0A1W4W882 A0A0Q9WNT8 A0A1W4WIJ3 A0A0Q5TGS1 A0A0Q5T381 A0A1W4VSN5 A0A0Q9VZ07 A0A1W4WJS5 A0A1W4VEP6 A0A1W4VEE2 A0A0Q9WAJ9 A0A1W4W883 A0A0T6AVH7 A0A0Q5TG56 Q9W4S9-2 A0A154P7D8 D0IQA0 K7J1X9 A0A1I8QD97 A0A1I8QDB2 B4IA32 A0A1B6E622 A0A0Q9WNR7 A0A151WH39 Q9W4S9 A0A1I8QD91 Q8MS17 T1PME5 H8F4P8 Q9W4T4 A0A1W4WJI5 A0A1W4WIJ8 A0A1B6DRH6 A0A1W4VRE9 A0A1W4VS49 A0A1W4WJJ1 A0A1W4W887 A0A1B6CIC6 A0A1B6CZC8 P12252-7 A0A232FBZ1 A0A1W4W888 M9PDN2 P12252-9 A0A1W4WJT5 C9QPC1 A0A0L0C0L2 Q8IRU4 A0A1W4W891 A0A0C9PRG4 B3N1U9 A0A0Q5T2H8 A0A195C111 P12252-8
A0A212ENZ6 A0A1B0CBX1 A0A2A3E5J0 V9I967 B4JMU1 A0A3B0K0B8 V9IAZ4 A0A0Q9W6Z7 A0A0Q9WB84 A0A0M4F9F4 A0A0Q9VZ20 A0A1I8QDE8 A0A1I8QD96 A0A1I8QD92 A0A1L8DLH8 A0A1B6ERJ9 A0A1I8QDE4 A0A1I8QDA1 A0A1A9ZLK7 A0A1L8DLL1 A0A0Q9WZA2 A0A0Q9WNL3 A0A1B6FHL2 A0A1L8DL56 P12252-5 A0A1I8MIT5 E2QD73 A0A0Q9VZ02 A0A1I8QD76 P12252 P12252-4 A0A0Q9VZB9 A0A1I8QDA8 B4MJA8 E2AWF4 A0A1L8DLD7 A0A195FWK9 B4MCU3 A0A0Q9WBB8 A0A1A9WIK4 A0A1W4W882 A0A0Q9WNT8 A0A1W4WIJ3 A0A0Q5TGS1 A0A0Q5T381 A0A1W4VSN5 A0A0Q9VZ07 A0A1W4WJS5 A0A1W4VEP6 A0A1W4VEE2 A0A0Q9WAJ9 A0A1W4W883 A0A0T6AVH7 A0A0Q5TG56 Q9W4S9-2 A0A154P7D8 D0IQA0 K7J1X9 A0A1I8QD97 A0A1I8QDB2 B4IA32 A0A1B6E622 A0A0Q9WNR7 A0A151WH39 Q9W4S9 A0A1I8QD91 Q8MS17 T1PME5 H8F4P8 Q9W4T4 A0A1W4WJI5 A0A1W4WIJ8 A0A1B6DRH6 A0A1W4VRE9 A0A1W4VS49 A0A1W4WJJ1 A0A1W4W887 A0A1B6CIC6 A0A1B6CZC8 P12252-7 A0A232FBZ1 A0A1W4W888 M9PDN2 P12252-9 A0A1W4WJT5 C9QPC1 A0A0L0C0L2 Q8IRU4 A0A1W4W891 A0A0C9PRG4 B3N1U9 A0A0Q5T2H8 A0A195C111 P12252-8
PDB
4X0F
E-value=7.0757e-163,
Score=1474
Ontologies
PATHWAY
GO
GO:0046872
GO:0004114
GO:0007165
GO:0040040
GO:0048149
GO:0010738
GO:0072375
GO:0019933
GO:0048675
GO:0007614
GO:0005634
GO:0007268
GO:0005829
GO:0007615
GO:0001661
GO:0006198
GO:0007612
GO:0046958
GO:0007613
GO:0007619
GO:0004115
GO:0007623
GO:0007617
GO:0045475
GO:0000003
GO:0008306
GO:0008355
GO:0007611
GO:0008081
GO:0003824
GO:0016829
GO:0005737
GO:0048869
GO:0003676
GO:0016491
GO:0015930
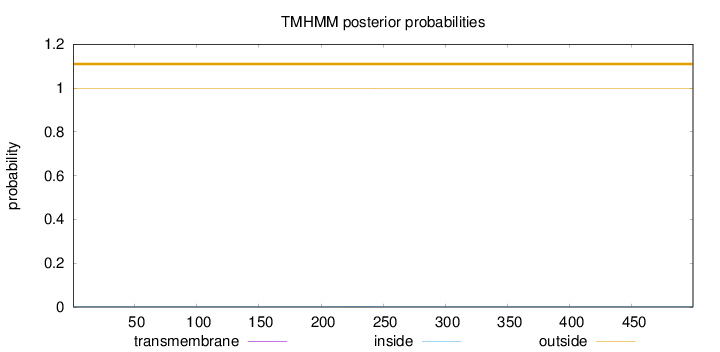
Topology
Length:
499
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00722
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00146
outside
1 - 499
Population Genetic Test Statistics
Pi
245.507637
Theta
185.100581
Tajima's D
0.700527
CLR
0.381695
CSRT
0.572821358932053
Interpretation
Uncertain
