Gene
KWMTBOMO13202
Annotation
PREDICTED:_serine/threonine-protein_kinase_Nek8-like_[Bombyx_mori]
Full name
Serine/threonine-protein kinase Nek8
Alternative Name
Never in mitosis A-related kinase 8
Nima-related protein kinase 12a
Nima-related protein kinase 12a
Location in the cell
PlasmaMembrane Reliability : 2.351
Sequence
CDS
ATGGATCTACAGAATCGTTTTACTAATCAAAAATTGTCAATATTGAAGACAATAGGCAAAGGAACTTACGGAAATGTGTACTTGTGCGAGAATCATGAAAACCATTTATTAACTATTGTAAAAGATATTCAACTGAACGTCAAAATACAACAACATGAACAGGACATCGCAAACGAAGTTAGAATTTTATCTACTATGAAACATCCCAACATCATTAGTTTCTACCAGTGCTTCTACGCCGAAGATTACGTAATGATATCAATGGAATACGCAACTTGTGGAAATTTAGCCGAGTTCATGTACCTTCGGTGCCCGCAAGTCATCAAACAAGAGGAAATTCTGTTCTACTTTTGCCAAATACTCTTAGGCGTAAGTTACATTCACAATTTGGACATCATTCACCGAGATTTGAAAGCAGAAAATATCTTACTGACTGGAAAATATGGTATAATAGTTAAAATAGGCGACTTTGGAATTTCAAAAATGTTAGCAAGTGCCAAGAAGACTTCAACAGTCATCGGAACACCTTACTACTTGGCGCCGGAACTATGCGAAGGTCAACCTTATGACACGAAAAGCGACATTTGGGCGCTCGGCTGTTTGCTTTACGAAATGTGTACACATAAACGTGCTTTTGAATCGGAGACTCTCGTCGGTCTAGTCAAAGCAATCACAAGTGGTAGCGTCCACCCTATCGACTTGTTTTTATACGACAGAGGGATGCAAGACCTTATTGATTCGATGCTGTCAATATTGCCAAACAAAAGGCCGACCATCAAAGAAGTTATGGGAAAAGAAATCATTTTACCCGTTATTTACACGGTTTATTTGGACGCGGGTGACGACGATTTATTGATTTTGAAAGCTAAAGAATTTATGTGA
Protein
MDLQNRFTNQKLSILKTIGKGTYGNVYLCENHENHLLTIVKDIQLNVKIQQHEQDIANEVRILSTMKHPNIISFYQCFYAEDYVMISMEYATCGNLAEFMYLRCPQVIKQEEILFYFCQILLGVSYIHNLDIIHRDLKAENILLTGKYGIIVKIGDFGISKMLASAKKTSTVIGTPYYLAPELCEGQPYDTKSDIWALGCLLYEMCTHKRAFESETLVGLVKAITSGSVHPIDLFLYDRGMQDLIDSMLSILPNKRPTIKEVMGKEIILPVIYTVYLDAGDDDLLILKAKEFM
Summary
Description
Required for renal tubular integrity. May regulate local cytoskeletal structure in kidney tubule epithelial cells. May regulate ciliary biogenesis through targeting of proteins to the cilia. Plays a role in organogenesis and is involved in the regulation of the Hippo signaling pathway (By similarity).
Required for renal tubular integrity. May regulate local cytoskeletal structure in kidney tubule epithelial cells. May regulate ciliary biogenesis through targeting of proteins to the cilia (By similarity). Plays a role in organogenesis and is involved in the regulation of the Hippo signaling pathway.
Required for renal tubular integrity. May regulate local cytoskeletal structure in kidney tubule epithelial cells. May regulate ciliary biogenesis through targeting of proteins to the cilia. Plays a role in organogenesis and is involved in the regulation of the Hippo signaling pathway.
Required for renal tubular integrity. May regulate local cytoskeletal structure in kidney tubule epithelial cells. May regulate ciliary biogenesis through targeting of proteins to the cilia (By similarity). Plays a role in organogenesis and is involved in the regulation of the Hippo signaling pathway.
Required for renal tubular integrity. May regulate local cytoskeletal structure in kidney tubule epithelial cells. May regulate ciliary biogenesis through targeting of proteins to the cilia. Plays a role in organogenesis and is involved in the regulation of the Hippo signaling pathway.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Subunit
Interacts with PKD2; may regulate PKD2 targeting to the cilium. Component of a complex containing at least ANKS6, INVS, NEK8 and NPHP3 (PubMed:23793029). ANKS6 may organize complex assembly by linking INVS and NPHP3 to NEK8 and INVS may target it to the proximal ciliary axoneme. Interacts with ANKS3 (By similarity).
Interacts with PKD2; may regulate PKD2 targeting to the cilium (By similarity). Component of a complex containing at least ANKS6, INVS, NEK8 and NPHP3. ANKS6 may organize complex assembly by linking INVS and NPHP3 to NEK8 and INVS may target the complex to the proximal ciliary axoneme. Interacts with ANKS3 (By similarity).
Interacts with PKD2; may regulate PKD2 targeting to the cilium. Component of a complex containing at least ANKS6, INVS, NEK8 and NPHP3. ANKS6 may organize complex assembly by linking INVS and NPHP3 to NEK8 and INVS may target the complex to the proximal ciliary axoneme (By similarity). Interacts with ANKS3 (PubMed:25671767).
Interacts with PKD2; may regulate PKD2 targeting to the cilium (By similarity). Component of a complex containing at least ANKS6, INVS, NEK8 and NPHP3. ANKS6 may organize complex assembly by linking INVS and NPHP3 to NEK8 and INVS may target the complex to the proximal ciliary axoneme. Interacts with ANKS3 (By similarity).
Interacts with PKD2; may regulate PKD2 targeting to the cilium. Component of a complex containing at least ANKS6, INVS, NEK8 and NPHP3. ANKS6 may organize complex assembly by linking INVS and NPHP3 to NEK8 and INVS may target the complex to the proximal ciliary axoneme (By similarity). Interacts with ANKS3 (PubMed:25671767).
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. NEK Ser/Thr protein kinase family. NIMA subfamily.
Belongs to the protein kinase superfamily. NEK Ser/Thr protein kinase family. NIMA subfamily.
Keywords
ATP-binding
Cell projection
Cilium
Complete proteome
Cytoplasm
Cytoskeleton
Disease mutation
Kinase
Magnesium
Metal-binding
Nucleotide-binding
Phosphoprotein
Reference proteome
Repeat
Transferase
Ciliopathy
Nephronophthisis
Feature
chain Serine/threonine-protein kinase Nek8
sequence variant In NPHP9; a full-length mouse NEK8 construct containing the mutation shows a defect in ciliary localization with no apparent effect on ciliation, mitosis or centriole number; dbSNP:rs199962228.
sequence variant In NPHP9; a full-length mouse NEK8 construct containing the mutation shows a defect in ciliary localization with no apparent effect on ciliation, mitosis or centriole number; dbSNP:rs199962228.
Uniprot
A0A212F1I3
A0A2A4J3N5
A0A194PY50
A0A3S2LCX9
A0A194R0U8
A0A0L7LCN4
+ More
A0A0L7K4C3 A0A1Y1MT33 J9NYR9 H9H1W0 A0A146YJB5 A0A2J8LIA4 K7EL04 A0A2J8TMM9 C3ZJ42 A0A3M0KIA4 B4DW95 H0Z713 A0A1S3H2T7 A0A1S3F541 A0A2R2MJK9 A0A1V4K2E2 A0A218V1J8 A0A2I0LKT7 A0A384CH41 E1BRZ2 A0A2Y9T5S2 A0A3L8S080 A0A1U7TXZ9 G3IGI2 A0A3Q7MJK5 A0A093GRV6 A0A226P183 A0A3L7GLT7 A0A226MWQ9 A0A2U3VW84 A0A2Y9E1J9 A0A2U3XJJ3 A0A341AR24 D3ZGQ5 M3X8S2 A0A3Q7T4X0 Q6DJA8 L5LKK7 A0A2U4ANQ4 A0A2K5DQ04 A0A2Y9JXW2 A0A1S3A7U2 A0A2Y9I7H5 A0A1W4ZSK4 A0A3P4M9Y8 G3TB80 M3YRU0 A0A2K6FSD9 A0A3Q7Y6N9 G3ULQ9 A0A2U4ANN3 G1QRK6 A0A096P3C4 A0A1W4ZJ40 E1B827 A0A2K6DDI2 A0A2K5XGK4 A0A2K5NPU5 G7NGK7 G7PTY4 A0A2K6P0B4 F6UM89 A0A2Y9Q216 W5P0W7 Q86SG6 A0A1L8H516 A0A093GNQ0 A0A091D5M3 G5BVI5 S7NAL6 Q91ZR4 A0A2K6LPL6 K1Q884 G9KD28 A0A2R8ZBY5 H2QCJ1 A0A1U7QY18 G1NW63 G1SE82 H0X674 A0A2J8TMR5 A0A2K6T597 H0UW90 F6YW35 A0A212D7H0 A0A093IYN3 A0A2Y9FFJ8 I3L8I2 A0A0P6J5G7 A0A341AUP5 A0A384A9J9 A0A2U4ANQ0
A0A0L7K4C3 A0A1Y1MT33 J9NYR9 H9H1W0 A0A146YJB5 A0A2J8LIA4 K7EL04 A0A2J8TMM9 C3ZJ42 A0A3M0KIA4 B4DW95 H0Z713 A0A1S3H2T7 A0A1S3F541 A0A2R2MJK9 A0A1V4K2E2 A0A218V1J8 A0A2I0LKT7 A0A384CH41 E1BRZ2 A0A2Y9T5S2 A0A3L8S080 A0A1U7TXZ9 G3IGI2 A0A3Q7MJK5 A0A093GRV6 A0A226P183 A0A3L7GLT7 A0A226MWQ9 A0A2U3VW84 A0A2Y9E1J9 A0A2U3XJJ3 A0A341AR24 D3ZGQ5 M3X8S2 A0A3Q7T4X0 Q6DJA8 L5LKK7 A0A2U4ANQ4 A0A2K5DQ04 A0A2Y9JXW2 A0A1S3A7U2 A0A2Y9I7H5 A0A1W4ZSK4 A0A3P4M9Y8 G3TB80 M3YRU0 A0A2K6FSD9 A0A3Q7Y6N9 G3ULQ9 A0A2U4ANN3 G1QRK6 A0A096P3C4 A0A1W4ZJ40 E1B827 A0A2K6DDI2 A0A2K5XGK4 A0A2K5NPU5 G7NGK7 G7PTY4 A0A2K6P0B4 F6UM89 A0A2Y9Q216 W5P0W7 Q86SG6 A0A1L8H516 A0A093GNQ0 A0A091D5M3 G5BVI5 S7NAL6 Q91ZR4 A0A2K6LPL6 K1Q884 G9KD28 A0A2R8ZBY5 H2QCJ1 A0A1U7QY18 G1NW63 G1SE82 H0X674 A0A2J8TMR5 A0A2K6T597 H0UW90 F6YW35 A0A212D7H0 A0A093IYN3 A0A2Y9FFJ8 I3L8I2 A0A0P6J5G7 A0A341AUP5 A0A384A9J9 A0A2U4ANQ0
EC Number
2.7.11.1
Pubmed
22118469
26354079
26227816
28004739
16341006
20838655
+ More
16625196 18563158 20360741 23371554 15592404 30282656 21804562 24621616 29704459 15057822 23793029 17975172 19393038 22002653 25362486 17431167 20809919 15489334 17974005 15019993 23418306 18199800 27762356 21993625 12421721 16141072 16267153 18235101 25671767 22992520 23236062 22722832 16136131 21993624 19892987
16625196 18563158 20360741 23371554 15592404 30282656 21804562 24621616 29704459 15057822 23793029 17975172 19393038 22002653 25362486 17431167 20809919 15489334 17974005 15019993 23418306 18199800 27762356 21993625 12421721 16141072 16267153 18235101 25671767 22992520 23236062 22722832 16136131 21993624 19892987
EMBL
AGBW02010868
OWR47598.1
NWSH01003294
PCG66601.1
KQ459586
KPI97953.1
+ More
RSAL01000040 RVE50949.1 KQ460883 KPJ11327.1 JTDY01001687 KOB73154.1 JTDY01010679 KOB58103.1 GEZM01024956 JAV87675.1 AAEX03006669 GCES01032070 JAR54253.1 NBAG03000289 PNI46994.1 AC010761 NDHI03003489 PNJ34275.1 GG666631 EEN47471.1 QRBI01000111 RMC10720.1 AK301429 BAG62957.1 ABQF01026366 LSYS01005108 OPJ78619.1 MUZQ01000070 OWK59905.1 AKCR02000235 PKK18046.1 AADN05000057 QUSF01000101 RLV92295.1 JH002576 EGW14552.1 KL216427 KFV69529.1 AWGT02000220 OXB73372.1 RAZU01001831 RLQ54628.1 MCFN01000384 OXB59540.1 AABR06064754 CH473948 AANG04004035 BC075274 AAH75274.1 KB110620 ELK26939.1 CYRY02007629 VCW76608.1 AEYP01021287 AEYP01021288 ADFV01099414 ADFV01099415 ADFV01099416 ADFV01099417 AHZZ02009949 CM001268 EHH24644.1 AQIA01027020 CM001291 EHH57833.1 JSUE03016317 AMGL01017759 AMGL01017760 AY242354 AY267371 CH471159 BC112240 BC113705 AL833909 CM004469 OCT91101.1 KL205569 KFV71933.1 KN123351 KFO25565.1 JH172092 EHB13321.1 KE163905 EPQ14334.1 AF407579 AK154358 BC070457 JH818702 EKC30153.1 JP014205 AES02803.1 AJFE02040461 AACZ04041502 PNI46993.1 AAPE02030648 AAGW02049845 AAQR03132776 AAQR03132777 AAQR03132778 PNJ34274.1 AAKN02033340 MKHE01000005 OWK14199.1 KK569907 KFW06778.1 AEMK02000080 GEBF01004791 JAN98841.1
RSAL01000040 RVE50949.1 KQ460883 KPJ11327.1 JTDY01001687 KOB73154.1 JTDY01010679 KOB58103.1 GEZM01024956 JAV87675.1 AAEX03006669 GCES01032070 JAR54253.1 NBAG03000289 PNI46994.1 AC010761 NDHI03003489 PNJ34275.1 GG666631 EEN47471.1 QRBI01000111 RMC10720.1 AK301429 BAG62957.1 ABQF01026366 LSYS01005108 OPJ78619.1 MUZQ01000070 OWK59905.1 AKCR02000235 PKK18046.1 AADN05000057 QUSF01000101 RLV92295.1 JH002576 EGW14552.1 KL216427 KFV69529.1 AWGT02000220 OXB73372.1 RAZU01001831 RLQ54628.1 MCFN01000384 OXB59540.1 AABR06064754 CH473948 AANG04004035 BC075274 AAH75274.1 KB110620 ELK26939.1 CYRY02007629 VCW76608.1 AEYP01021287 AEYP01021288 ADFV01099414 ADFV01099415 ADFV01099416 ADFV01099417 AHZZ02009949 CM001268 EHH24644.1 AQIA01027020 CM001291 EHH57833.1 JSUE03016317 AMGL01017759 AMGL01017760 AY242354 AY267371 CH471159 BC112240 BC113705 AL833909 CM004469 OCT91101.1 KL205569 KFV71933.1 KN123351 KFO25565.1 JH172092 EHB13321.1 KE163905 EPQ14334.1 AF407579 AK154358 BC070457 JH818702 EKC30153.1 JP014205 AES02803.1 AJFE02040461 AACZ04041502 PNI46993.1 AAPE02030648 AAGW02049845 AAQR03132776 AAQR03132777 AAQR03132778 PNJ34274.1 AAKN02033340 MKHE01000005 OWK14199.1 KK569907 KFW06778.1 AEMK02000080 GEBF01004791 JAN98841.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000283053
UP000053240
UP000037510
+ More
UP000002254 UP000001645 UP000005640 UP000001554 UP000269221 UP000007754 UP000085678 UP000081671 UP000190648 UP000197619 UP000053872 UP000261680 UP000000539 UP000248484 UP000276834 UP000189704 UP000001075 UP000286641 UP000053875 UP000198419 UP000273346 UP000198323 UP000245340 UP000248480 UP000245341 UP000252040 UP000002494 UP000011712 UP000286640 UP000245320 UP000233020 UP000248482 UP000079721 UP000248481 UP000192224 UP000007646 UP000000715 UP000233160 UP000286642 UP000001073 UP000028761 UP000009136 UP000233120 UP000233140 UP000233060 UP000009130 UP000233100 UP000233200 UP000006718 UP000248483 UP000002356 UP000186698 UP000053584 UP000028990 UP000006813 UP000000589 UP000233180 UP000005408 UP000240080 UP000002277 UP000189706 UP000001074 UP000001811 UP000005225 UP000233220 UP000005447 UP000002281 UP000008227 UP000261681
UP000002254 UP000001645 UP000005640 UP000001554 UP000269221 UP000007754 UP000085678 UP000081671 UP000190648 UP000197619 UP000053872 UP000261680 UP000000539 UP000248484 UP000276834 UP000189704 UP000001075 UP000286641 UP000053875 UP000198419 UP000273346 UP000198323 UP000245340 UP000248480 UP000245341 UP000252040 UP000002494 UP000011712 UP000286640 UP000245320 UP000233020 UP000248482 UP000079721 UP000248481 UP000192224 UP000007646 UP000000715 UP000233160 UP000286642 UP000001073 UP000028761 UP000009136 UP000233120 UP000233140 UP000233060 UP000009130 UP000233100 UP000233200 UP000006718 UP000248483 UP000002356 UP000186698 UP000053584 UP000028990 UP000006813 UP000000589 UP000233180 UP000005408 UP000240080 UP000002277 UP000189706 UP000001074 UP000001811 UP000005225 UP000233220 UP000005447 UP000002281 UP000008227 UP000261681
Interpro
IPR017441
Protein_kinase_ATP_BS
+ More
IPR008271 Ser/Thr_kinase_AS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR000408 Reg_chr_condens
IPR009091 RCC1/BLIP-II
IPR001293 Znf_TRAF
IPR013083 Znf_RING/FYVE/PHD
IPR037307 TRAF4_MATH
IPR008974 TRAF-like
IPR002083 MATH/TRAF_dom
IPR008271 Ser/Thr_kinase_AS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR000408 Reg_chr_condens
IPR009091 RCC1/BLIP-II
IPR001293 Znf_TRAF
IPR013083 Znf_RING/FYVE/PHD
IPR037307 TRAF4_MATH
IPR008974 TRAF-like
IPR002083 MATH/TRAF_dom
Gene 3D
ProteinModelPortal
A0A212F1I3
A0A2A4J3N5
A0A194PY50
A0A3S2LCX9
A0A194R0U8
A0A0L7LCN4
+ More
A0A0L7K4C3 A0A1Y1MT33 J9NYR9 H9H1W0 A0A146YJB5 A0A2J8LIA4 K7EL04 A0A2J8TMM9 C3ZJ42 A0A3M0KIA4 B4DW95 H0Z713 A0A1S3H2T7 A0A1S3F541 A0A2R2MJK9 A0A1V4K2E2 A0A218V1J8 A0A2I0LKT7 A0A384CH41 E1BRZ2 A0A2Y9T5S2 A0A3L8S080 A0A1U7TXZ9 G3IGI2 A0A3Q7MJK5 A0A093GRV6 A0A226P183 A0A3L7GLT7 A0A226MWQ9 A0A2U3VW84 A0A2Y9E1J9 A0A2U3XJJ3 A0A341AR24 D3ZGQ5 M3X8S2 A0A3Q7T4X0 Q6DJA8 L5LKK7 A0A2U4ANQ4 A0A2K5DQ04 A0A2Y9JXW2 A0A1S3A7U2 A0A2Y9I7H5 A0A1W4ZSK4 A0A3P4M9Y8 G3TB80 M3YRU0 A0A2K6FSD9 A0A3Q7Y6N9 G3ULQ9 A0A2U4ANN3 G1QRK6 A0A096P3C4 A0A1W4ZJ40 E1B827 A0A2K6DDI2 A0A2K5XGK4 A0A2K5NPU5 G7NGK7 G7PTY4 A0A2K6P0B4 F6UM89 A0A2Y9Q216 W5P0W7 Q86SG6 A0A1L8H516 A0A093GNQ0 A0A091D5M3 G5BVI5 S7NAL6 Q91ZR4 A0A2K6LPL6 K1Q884 G9KD28 A0A2R8ZBY5 H2QCJ1 A0A1U7QY18 G1NW63 G1SE82 H0X674 A0A2J8TMR5 A0A2K6T597 H0UW90 F6YW35 A0A212D7H0 A0A093IYN3 A0A2Y9FFJ8 I3L8I2 A0A0P6J5G7 A0A341AUP5 A0A384A9J9 A0A2U4ANQ0
A0A0L7K4C3 A0A1Y1MT33 J9NYR9 H9H1W0 A0A146YJB5 A0A2J8LIA4 K7EL04 A0A2J8TMM9 C3ZJ42 A0A3M0KIA4 B4DW95 H0Z713 A0A1S3H2T7 A0A1S3F541 A0A2R2MJK9 A0A1V4K2E2 A0A218V1J8 A0A2I0LKT7 A0A384CH41 E1BRZ2 A0A2Y9T5S2 A0A3L8S080 A0A1U7TXZ9 G3IGI2 A0A3Q7MJK5 A0A093GRV6 A0A226P183 A0A3L7GLT7 A0A226MWQ9 A0A2U3VW84 A0A2Y9E1J9 A0A2U3XJJ3 A0A341AR24 D3ZGQ5 M3X8S2 A0A3Q7T4X0 Q6DJA8 L5LKK7 A0A2U4ANQ4 A0A2K5DQ04 A0A2Y9JXW2 A0A1S3A7U2 A0A2Y9I7H5 A0A1W4ZSK4 A0A3P4M9Y8 G3TB80 M3YRU0 A0A2K6FSD9 A0A3Q7Y6N9 G3ULQ9 A0A2U4ANN3 G1QRK6 A0A096P3C4 A0A1W4ZJ40 E1B827 A0A2K6DDI2 A0A2K5XGK4 A0A2K5NPU5 G7NGK7 G7PTY4 A0A2K6P0B4 F6UM89 A0A2Y9Q216 W5P0W7 Q86SG6 A0A1L8H516 A0A093GNQ0 A0A091D5M3 G5BVI5 S7NAL6 Q91ZR4 A0A2K6LPL6 K1Q884 G9KD28 A0A2R8ZBY5 H2QCJ1 A0A1U7QY18 G1NW63 G1SE82 H0X674 A0A2J8TMR5 A0A2K6T597 H0UW90 F6YW35 A0A212D7H0 A0A093IYN3 A0A2Y9FFJ8 I3L8I2 A0A0P6J5G7 A0A341AUP5 A0A384A9J9 A0A2U4ANQ0
PDB
4B9D
E-value=1.08629e-48,
Score=486
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Predominantly cytoplasmic. Localizes to the proximal region of the primary cilium and is not observed in dividing cells (By similarity). With evidence from 1 publications.
Cytoskeleton Predominantly cytoplasmic. Localizes to the proximal region of the primary cilium and is not observed in dividing cells (By similarity). With evidence from 1 publications.
Cell projection Predominantly cytoplasmic. Localizes to the proximal region of the primary cilium and is not observed in dividing cells (By similarity). With evidence from 1 publications.
Cilium Predominantly cytoplasmic. Localizes to the proximal region of the primary cilium and is not observed in dividing cells (By similarity). With evidence from 1 publications.
Cytoskeleton Predominantly cytoplasmic. Localizes to the proximal region of the primary cilium and is not observed in dividing cells (By similarity). With evidence from 1 publications.
Cell projection Predominantly cytoplasmic. Localizes to the proximal region of the primary cilium and is not observed in dividing cells (By similarity). With evidence from 1 publications.
Cilium Predominantly cytoplasmic. Localizes to the proximal region of the primary cilium and is not observed in dividing cells (By similarity). With evidence from 1 publications.
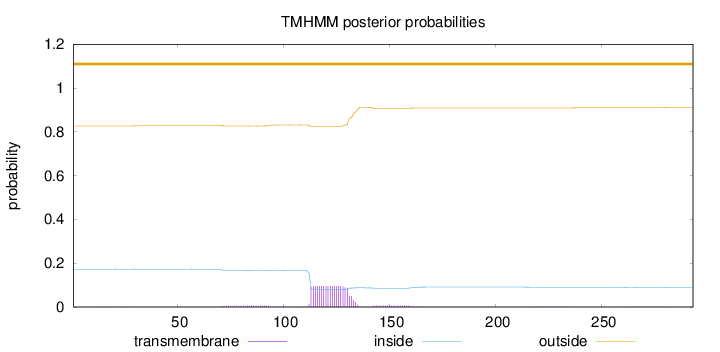
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.22931
Exp number, first 60 AAs:
0.00362
Total prob of N-in:
0.17294
outside
1 - 293
Population Genetic Test Statistics
Pi
231.523973
Theta
162.797981
Tajima's D
2.670674
CLR
0
CSRT
0.95865206739663
Interpretation
Uncertain
