Pre Gene Modal
BGIBMGA009955
Annotation
PREDICTED:_deoxyribose-phosphate_aldolase-like_isoform_X1_[Amyelois_transitella]
Full name
Deoxyribose-phosphate aldolase
Alternative Name
2-deoxy-D-ribose 5-phosphate aldolase
Phosphodeoxyriboaldolase
Phosphodeoxyriboaldolase
Location in the cell
PlasmaMembrane Reliability : 1.801
Sequence
CDS
ATGGTGCGAGCTATTTTTGAAACTATTGATGATATGCCCTTAGATAATACATTTATAAATAAAAGCAATGTAGATTCTCAAGTAAAATCAATATTATACAACAATGTTGTGTCTCCTAAGGATGAGAGGGCATGGCTTATGAAAGCAATCACTCTAATTGACTTGACAACACTTTCTGGTGATGACACATCATCAAATGTTAGAAGACTTTGTCGGAAGGCTGCATATCCAATTCCAATTGATATTATGAATCCTTCTGATAATCTTCACACTGCAGCTGTCTGTGTCTACCCTACAAAGGTGCCTGATGTATGTACTGCTCTGAAGGCTATGAAGTTGGAAAAAAACATACAAGTGGCTGCAGTGGCTACAGGATTTCCGTCTGGTTTGTATCCTCTCGCAACGAGATTAGAAGAGATTCGGATGACCGTAGGGTATGGTGCAACAGAAATTGATGTGGTTATCGATAGGAGTTTAGTTTTGACCGGAGAATGGGAGACACTGTTTAATGAAATACAACAAATGAAGAAAGCATGCGGATCTGCACATCTCAAAGTTATTCTTGGCGTTGGCGAATTGGGAACTTATTATAATGTGTACAAAGCTTCAATGATCGCCATGATGGCTGGAGCGGACTTCATTAAGACATCGACCGGCAAAGAAGCCGTAAACGCAACGCTACCCATTGGTCTTGTCATGTGTCGGGCTATTAGAAGATATTATCAGATGACCGGTCATAAGGTTGGACTTAAACCGGCAGGTGGAATAAAAACTGTTCAGGATACACTGAATTGGTTAGTTCTTTTGTACACAGAGCTGGGTCCTGACTGGCTCACACCTGCCCTATTCAGGATTGGAGCGTCCAGTCTTCTGGACGTGATTATAAATCATTTAAAATCCACAAATCAGATTAGCTGTAGCTGA
Protein
MVRAIFETIDDMPLDNTFINKSNVDSQVKSILYNNVVSPKDERAWLMKAITLIDLTTLSGDDTSSNVRRLCRKAAYPIPIDIMNPSDNLHTAAVCVYPTKVPDVCTALKAMKLEKNIQVAAVATGFPSGLYPLATRLEEIRMTVGYGATEIDVVIDRSLVLTGEWETLFNEIQQMKKACGSAHLKVILGVGELGTYYNVYKASMIAMMAGADFIKTSTGKEAVNATLPIGLVMCRAIRRYYQMTGHKVGLKPAGGIKTVQDTLNWLVLLYTELGPDWLTPALFRIGASSLLDVIINHLKSTNQISCS
Summary
Description
Catalyzes a reversible aldol reaction between acetaldehyde and D-glyceraldehyde 3-phosphate to generate 2-deoxy-D-ribose 5-phosphate. Participates in stress granule (SG) assembly. May allow ATP production from extracellular deoxyinosine in conditions of energy deprivation.
Catalytic Activity
2-deoxy-D-ribose 5-phosphate = acetaldehyde + D-glyceraldehyde 3-phosphate
Subunit
Interacts with YBX1.
Similarity
Belongs to the DeoC/FbaB aldolase family. DeoC type 2 subfamily.
Keywords
Complete proteome
Cytoplasm
Lyase
Nucleus
Reference proteome
Schiff base
Feature
chain Deoxyribose-phosphate aldolase
Uniprot
H9JKA4
A0A2H1VAU7
A0A2H1V8V4
A0A3S2LJY7
S4PMF7
A0A194PYX5
+ More
A0A212F1L4 A0A1Q3FDU9 B0WRC7 A0A2P8XFX9 A0A182F0W7 W5JAU3 A0A2M4BUS5 T1DKA8 D7EJ08 A0A2M4AV24 A0A2M4AVK4 A0A2J7RJI0 A0A182NF31 Q17G09 A0A182G2I8 A0A182QLF1 G1LA40 M3YJ84 A0A3Q2HI48 G1T6C1 A0A0V0G982 A0A2Y9RUV1 A0A182YCH6 A0A2I2UHA8 A0A2I3SJ20 A0A3P8WFT0 I3LXP1 A0A2I2ZSG6 V9KVM3 A0A1S4F4A5 A0A099Z5N7 A0A182M158 A0A3B4XJT4 A0A2R8ZRB1 H2Q5J5 A0A3Q7R5E2 Q9Y315 A0A2I3T1B0 F1PPB6 A0A2K5C0A3 A0A2K5C097 A0A2K5C0C1 A0A2K6LDR6 A0A3P8WDP3 A0A2K6T142 A0A2D0T4E5 A0A2K6T1C1 A0A2R8ZUH6 H9F930 A0A3Q2IDE8 D2HA66 G3QQS5 A0A2R8ZRC3 A0A2I3SNI6 A0A2U3WEM0 A0A2K6T1C5 G7PJY8 A0A182R6K4 G3RZY7 A0A2K6ED78 A0A2K5U9U9 A0A2K5XZ81 A0A0D9R7I1 A0A2I3N060 A0A2K5MA98 A0A2K5J1C3 F6XLA4 A0A2Y9H3J4 A0A1U7S877 A0A3B4V3P9 A0A2K5C0E7 A0A2K6LDS3 A0A3Q7VAG0 W5KVU7 A0A2K5J179 A0A384DEQ6 A0A2K6ED76 A0A2K5U9U0 A0A2K5XZ94 A0A2I3LLF2 A0A2K5MA88 A0A2K5J0W7 A0A2K5U9S7 A0A096N672 I3K9K7 M1EKL9 A0A2K5XZ73 A0A2I3NHF5 A0A2Y9IU31 H0X8G9 A0A182WH53 A0A2U9BB25 A0A2K6T1F6 A0A2U3ZGX2
A0A212F1L4 A0A1Q3FDU9 B0WRC7 A0A2P8XFX9 A0A182F0W7 W5JAU3 A0A2M4BUS5 T1DKA8 D7EJ08 A0A2M4AV24 A0A2M4AVK4 A0A2J7RJI0 A0A182NF31 Q17G09 A0A182G2I8 A0A182QLF1 G1LA40 M3YJ84 A0A3Q2HI48 G1T6C1 A0A0V0G982 A0A2Y9RUV1 A0A182YCH6 A0A2I2UHA8 A0A2I3SJ20 A0A3P8WFT0 I3LXP1 A0A2I2ZSG6 V9KVM3 A0A1S4F4A5 A0A099Z5N7 A0A182M158 A0A3B4XJT4 A0A2R8ZRB1 H2Q5J5 A0A3Q7R5E2 Q9Y315 A0A2I3T1B0 F1PPB6 A0A2K5C0A3 A0A2K5C097 A0A2K5C0C1 A0A2K6LDR6 A0A3P8WDP3 A0A2K6T142 A0A2D0T4E5 A0A2K6T1C1 A0A2R8ZUH6 H9F930 A0A3Q2IDE8 D2HA66 G3QQS5 A0A2R8ZRC3 A0A2I3SNI6 A0A2U3WEM0 A0A2K6T1C5 G7PJY8 A0A182R6K4 G3RZY7 A0A2K6ED78 A0A2K5U9U9 A0A2K5XZ81 A0A0D9R7I1 A0A2I3N060 A0A2K5MA98 A0A2K5J1C3 F6XLA4 A0A2Y9H3J4 A0A1U7S877 A0A3B4V3P9 A0A2K5C0E7 A0A2K6LDS3 A0A3Q7VAG0 W5KVU7 A0A2K5J179 A0A384DEQ6 A0A2K6ED76 A0A2K5U9U0 A0A2K5XZ94 A0A2I3LLF2 A0A2K5MA88 A0A2K5J0W7 A0A2K5U9S7 A0A096N672 I3K9K7 M1EKL9 A0A2K5XZ73 A0A2I3NHF5 A0A2Y9IU31 H0X8G9 A0A182WH53 A0A2U9BB25 A0A2K6T1F6 A0A2U3ZGX2
EC Number
4.1.2.4
Pubmed
19121390
23622113
26354079
22118469
29403074
20920257
+ More
23761445 18362917 19820115 17510324 26483478 20010809 19892987 21993624 25244985 17975172 16136131 24487278 22398555 24402279 22722832 10810093 15489334 21269460 25229427 24275569 16341006 25319552 22002653 17431167 25329095 25186727 23236062
23761445 18362917 19820115 17510324 26483478 20010809 19892987 21993624 25244985 17975172 16136131 24487278 22398555 24402279 22722832 10810093 15489334 21269460 25229427 24275569 16341006 25319552 22002653 17431167 25329095 25186727 23236062
EMBL
BABH01036839
ODYU01001568
SOQ37965.1
ODYU01001280
SOQ37280.1
RSAL01000086
+ More
RVE48313.1 GAIX01003575 JAA88985.1 KQ459586 KPI97949.1 AGBW02010868 OWR47603.1 GFDL01009377 JAV25668.1 DS232054 EDS33317.1 PYGN01002284 PSN30911.1 ADMH02001915 ETN60523.1 GGFJ01007668 MBW56809.1 GAMD01001036 JAB00555.1 DS497692 EFA12496.1 GGFK01011316 MBW44637.1 GGFK01011483 MBW44804.1 NEVH01002992 PNF40983.1 CH477267 EAT45461.1 JXUM01039798 KQ561218 KXJ79315.1 AXCN02000830 ACTA01009862 ACTA01017862 ACTA01025862 AEYP01039891 AEYP01039892 AEYP01039893 AEYP01039894 AAGW02044803 AAGW02044804 AAGW02044805 GECL01001608 JAP04516.1 AANG04001751 AACZ04031281 AGTP01068389 AGTP01068390 AGTP01068391 AGTP01068392 CABD030083016 CABD030083017 CABD030083018 CABD030083019 CABD030083020 CABD030083021 JW870172 AFP02690.1 KL889661 KGL77016.1 AXCM01002218 AJFE02056092 AJFE02056093 AJFE02056094 GABC01009645 GABF01005996 GABD01005783 GABE01004242 NBAG03000025 JAA01693.1 JAA16149.1 JAA27317.1 JAA40497.1 PNI98334.1 AF132960 AK222541 BC056234 AAEX03015236 JU327383 AFE71139.1 GL192621 EFB27991.1 CM001286 EHH66123.1 AQIA01010186 AQIA01010187 AQIA01010188 AQIB01128531 AHZZ02001714 AHZZ02001715 JSUE03006235 JSUE03006236 JSUE03006237 JSUE03006238 JU471140 JV044385 CM001263 AFH27944.1 AFI34456.1 EHH20554.1 AERX01038186 JP008557 AER97154.1 AAQR03139537 AAQR03139538 AAQR03139539 CP026246 AWP00952.1
RVE48313.1 GAIX01003575 JAA88985.1 KQ459586 KPI97949.1 AGBW02010868 OWR47603.1 GFDL01009377 JAV25668.1 DS232054 EDS33317.1 PYGN01002284 PSN30911.1 ADMH02001915 ETN60523.1 GGFJ01007668 MBW56809.1 GAMD01001036 JAB00555.1 DS497692 EFA12496.1 GGFK01011316 MBW44637.1 GGFK01011483 MBW44804.1 NEVH01002992 PNF40983.1 CH477267 EAT45461.1 JXUM01039798 KQ561218 KXJ79315.1 AXCN02000830 ACTA01009862 ACTA01017862 ACTA01025862 AEYP01039891 AEYP01039892 AEYP01039893 AEYP01039894 AAGW02044803 AAGW02044804 AAGW02044805 GECL01001608 JAP04516.1 AANG04001751 AACZ04031281 AGTP01068389 AGTP01068390 AGTP01068391 AGTP01068392 CABD030083016 CABD030083017 CABD030083018 CABD030083019 CABD030083020 CABD030083021 JW870172 AFP02690.1 KL889661 KGL77016.1 AXCM01002218 AJFE02056092 AJFE02056093 AJFE02056094 GABC01009645 GABF01005996 GABD01005783 GABE01004242 NBAG03000025 JAA01693.1 JAA16149.1 JAA27317.1 JAA40497.1 PNI98334.1 AF132960 AK222541 BC056234 AAEX03015236 JU327383 AFE71139.1 GL192621 EFB27991.1 CM001286 EHH66123.1 AQIA01010186 AQIA01010187 AQIA01010188 AQIB01128531 AHZZ02001714 AHZZ02001715 JSUE03006235 JSUE03006236 JSUE03006237 JSUE03006238 JU471140 JV044385 CM001263 AFH27944.1 AFI34456.1 EHH20554.1 AERX01038186 JP008557 AER97154.1 AAQR03139537 AAQR03139538 AAQR03139539 CP026246 AWP00952.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000002320
UP000245037
+ More
UP000069272 UP000000673 UP000007266 UP000235965 UP000075884 UP000008820 UP000069940 UP000249989 UP000075886 UP000008912 UP000000715 UP000002281 UP000001811 UP000248480 UP000076408 UP000011712 UP000002277 UP000265120 UP000005215 UP000001519 UP000053641 UP000075883 UP000261360 UP000240080 UP000286641 UP000005640 UP000002254 UP000233020 UP000233180 UP000233220 UP000221080 UP000245340 UP000009130 UP000075900 UP000233120 UP000233100 UP000233140 UP000029965 UP000028761 UP000233060 UP000233080 UP000006718 UP000248481 UP000189705 UP000261420 UP000286642 UP000018467 UP000261680 UP000005207 UP000248482 UP000005225 UP000075920 UP000246464
UP000069272 UP000000673 UP000007266 UP000235965 UP000075884 UP000008820 UP000069940 UP000249989 UP000075886 UP000008912 UP000000715 UP000002281 UP000001811 UP000248480 UP000076408 UP000011712 UP000002277 UP000265120 UP000005215 UP000001519 UP000053641 UP000075883 UP000261360 UP000240080 UP000286641 UP000005640 UP000002254 UP000233020 UP000233180 UP000233220 UP000221080 UP000245340 UP000009130 UP000075900 UP000233120 UP000233100 UP000233140 UP000029965 UP000028761 UP000233060 UP000233080 UP000006718 UP000248481 UP000189705 UP000261420 UP000286642 UP000018467 UP000261680 UP000005207 UP000248482 UP000005225 UP000075920 UP000246464
Pfam
PF01791 DeoC
Gene 3D
CDD
ProteinModelPortal
H9JKA4
A0A2H1VAU7
A0A2H1V8V4
A0A3S2LJY7
S4PMF7
A0A194PYX5
+ More
A0A212F1L4 A0A1Q3FDU9 B0WRC7 A0A2P8XFX9 A0A182F0W7 W5JAU3 A0A2M4BUS5 T1DKA8 D7EJ08 A0A2M4AV24 A0A2M4AVK4 A0A2J7RJI0 A0A182NF31 Q17G09 A0A182G2I8 A0A182QLF1 G1LA40 M3YJ84 A0A3Q2HI48 G1T6C1 A0A0V0G982 A0A2Y9RUV1 A0A182YCH6 A0A2I2UHA8 A0A2I3SJ20 A0A3P8WFT0 I3LXP1 A0A2I2ZSG6 V9KVM3 A0A1S4F4A5 A0A099Z5N7 A0A182M158 A0A3B4XJT4 A0A2R8ZRB1 H2Q5J5 A0A3Q7R5E2 Q9Y315 A0A2I3T1B0 F1PPB6 A0A2K5C0A3 A0A2K5C097 A0A2K5C0C1 A0A2K6LDR6 A0A3P8WDP3 A0A2K6T142 A0A2D0T4E5 A0A2K6T1C1 A0A2R8ZUH6 H9F930 A0A3Q2IDE8 D2HA66 G3QQS5 A0A2R8ZRC3 A0A2I3SNI6 A0A2U3WEM0 A0A2K6T1C5 G7PJY8 A0A182R6K4 G3RZY7 A0A2K6ED78 A0A2K5U9U9 A0A2K5XZ81 A0A0D9R7I1 A0A2I3N060 A0A2K5MA98 A0A2K5J1C3 F6XLA4 A0A2Y9H3J4 A0A1U7S877 A0A3B4V3P9 A0A2K5C0E7 A0A2K6LDS3 A0A3Q7VAG0 W5KVU7 A0A2K5J179 A0A384DEQ6 A0A2K6ED76 A0A2K5U9U0 A0A2K5XZ94 A0A2I3LLF2 A0A2K5MA88 A0A2K5J0W7 A0A2K5U9S7 A0A096N672 I3K9K7 M1EKL9 A0A2K5XZ73 A0A2I3NHF5 A0A2Y9IU31 H0X8G9 A0A182WH53 A0A2U9BB25 A0A2K6T1F6 A0A2U3ZGX2
A0A212F1L4 A0A1Q3FDU9 B0WRC7 A0A2P8XFX9 A0A182F0W7 W5JAU3 A0A2M4BUS5 T1DKA8 D7EJ08 A0A2M4AV24 A0A2M4AVK4 A0A2J7RJI0 A0A182NF31 Q17G09 A0A182G2I8 A0A182QLF1 G1LA40 M3YJ84 A0A3Q2HI48 G1T6C1 A0A0V0G982 A0A2Y9RUV1 A0A182YCH6 A0A2I2UHA8 A0A2I3SJ20 A0A3P8WFT0 I3LXP1 A0A2I2ZSG6 V9KVM3 A0A1S4F4A5 A0A099Z5N7 A0A182M158 A0A3B4XJT4 A0A2R8ZRB1 H2Q5J5 A0A3Q7R5E2 Q9Y315 A0A2I3T1B0 F1PPB6 A0A2K5C0A3 A0A2K5C097 A0A2K5C0C1 A0A2K6LDR6 A0A3P8WDP3 A0A2K6T142 A0A2D0T4E5 A0A2K6T1C1 A0A2R8ZUH6 H9F930 A0A3Q2IDE8 D2HA66 G3QQS5 A0A2R8ZRC3 A0A2I3SNI6 A0A2U3WEM0 A0A2K6T1C5 G7PJY8 A0A182R6K4 G3RZY7 A0A2K6ED78 A0A2K5U9U9 A0A2K5XZ81 A0A0D9R7I1 A0A2I3N060 A0A2K5MA98 A0A2K5J1C3 F6XLA4 A0A2Y9H3J4 A0A1U7S877 A0A3B4V3P9 A0A2K5C0E7 A0A2K6LDS3 A0A3Q7VAG0 W5KVU7 A0A2K5J179 A0A384DEQ6 A0A2K6ED76 A0A2K5U9U0 A0A2K5XZ94 A0A2I3LLF2 A0A2K5MA88 A0A2K5J0W7 A0A2K5U9S7 A0A096N672 I3K9K7 M1EKL9 A0A2K5XZ73 A0A2I3NHF5 A0A2Y9IU31 H0X8G9 A0A182WH53 A0A2U9BB25 A0A2K6T1F6 A0A2U3ZGX2
PDB
5C6M
E-value=1.2485e-36,
Score=383
Ontologies
PATHWAY
GO
PANTHER
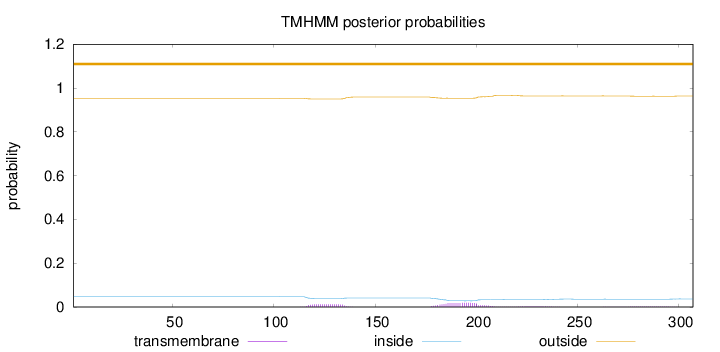
Topology
Subcellular location
Cytoplasm
Recruited to stress granules but not to processing bodies upon arsenite or clotrimazole treatment or energy deprivation. With evidence from 2 publications.
Cytoplasmic granule Recruited to stress granules but not to processing bodies upon arsenite or clotrimazole treatment or energy deprivation. With evidence from 2 publications.
Nucleus Recruited to stress granules but not to processing bodies upon arsenite or clotrimazole treatment or energy deprivation. With evidence from 2 publications.
Cytoplasmic granule Recruited to stress granules but not to processing bodies upon arsenite or clotrimazole treatment or energy deprivation. With evidence from 2 publications.
Nucleus Recruited to stress granules but not to processing bodies upon arsenite or clotrimazole treatment or energy deprivation. With evidence from 2 publications.
Length:
307
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.72198
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.04715
outside
1 - 307
Population Genetic Test Statistics
Pi
319.696009
Theta
230.889609
Tajima's D
1.583577
CLR
0
CSRT
0.806109694515274
Interpretation
Uncertain
