Gene
KWMTBOMO13194
Annotation
reverse_transcriptase_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.512
Sequence
CDS
ATGGTGACCAGGGGCCCCGGGATCGTCGCGGTACAGTTAGAAAATATCGTCGTGATCGGGGTGTACTTTTCTCCGAATCGGCCTACCGTCGAGTTCGAGCGGTTCCTGGACGGGCTAGAGGTGATCGCTCGTCACCTCGCTCCCCGTTCAGTGATACTGGCGGGGGACTTCAATGCGAAGTCTGTCGCTTGGGGTTCCCCATTCACGGACGCTCGTGGTAGGCTGCTGGAGGAGTGGGCGGTCGCGGTCGGTCTCTGTATCGTTAATAGGGGCTCGATCGCGACTTGCGTGCGGTGGACGGGCGAGTCTATCGTGGACTTGACGTTCGCGAGCTCGCCCGTCGCACGGCGCATCCTCGGCTGGAGAGTGGTGGTGGGGGCGGAATCGTTGTCCGATCACCGGTATATCCGGTACGATCTTTCTGCCCCTGCGCCGACGCCGGCTGCAGGTGCCCGCGGGGTTGACCCCCCGCAGAGTGCATCCCGGTCATTCCCTAGGTGGGCACTGAAGCTCCTGAACAGGGAGCTCCTGGTGGAGGCCTCTATGGTGGCGGCGTGGGCGCCCAAGCGTCCACAACCTGTCGATGTGGAAACCGAGGCCGAATGGTTCCGGGGCGCGATGCACCGCATATGTGATGCCTCGATGCCCCGGGTCGGCTCTCGGCACTCTGGACGGCGACAGTCGCCCTGGTGGTCGCCCGAAATTGCAAGACTGCGTGCGGTCTCCATTCGGGCGAGCCGCCGGTGCACTAGACACCGCCGTCGCCGCCGCCTGCGGCGCGACGAGCCCGTCGCGTTCGCGGAGGAGGAAGCTCGGCTGCACCGCGAATATCGCGCTGCGAAGGACGCCCTGCGGCTGGCCATCAGGCGGGCCAAGGAGCAGAACATGGAGAGACTCTTGGAGGCGCTCGAAGCGGATCCGTGGGGGAGCCCATACAAATTGGTACGCGGCAAGTTGCGCCCGTGGGCCCCCTCTATGACTGAGCGTCTCCAGCCCCAGCAGCTGCGGGACATCGTTTCGGCGTTGTTCCCGCAGGAGCGGGAGGGTTTTGTCCCTCCCGCTATGGGCGGGCCGTCTGACACCGTCGACGGCCAAGCTCCTGCTGAGGTGCCCCGTATTACGGAGGCAGAGCTCCGGGTGGCCGTTGTCAAGATGACTGCGAAAGACACGGCCCCCGGCCCGGACGGAGTCCACGGCCGGGTATTGGCCTTGGCCCTCGGTGCCCTGGGGGACCGGCTCCTTGAGCTATATAATGGCTGCTTGGAGTCGGGACGGTTTCCGTCGCTCTGGCGGACGGGTAGACTTGTGTTGTTGAGGAAGGAGGGGCGCCCGGTGGATACAGCCGCCGGATATCGTCCTATCGTGCTGCTGGACGAGGCGGGAAAATTGCTGGAACGTATTCTGGCTGCCCGCATCGTTCGGCACCTGGTCGGGGTGGGGCCTGACCTGTCGGCGGAGCAGTACGGCTTCCGGGAGGGCCGTTCGACCGTGGATGCAATTCTTCGCGTGCGGTCCCTCTCGGACGAGGCCGTCTCTCGGGGTGGGGTGGCGCTGGCGGTGTCTCTTGACATCGCCAACGCATTTAACACTCTGCCCTGGACCGTGATAGGGGGGGCACTGGAGAGGCATGGAGTGCCCCTCTACCTCCGCCGGCTGGTTGGGTCCTATTTGGGGGCCAGGTCGGTCGTATGTACCGGGTACGGTGGGACCCTTCATCGCTTTCCGGTCGTGCGTGGTGTTCCGCAGGGGTCGGTTCTCGGCCCCCTTTTGTGGAATATCGGGTACGACTGGGTGCTGAGGGGCGCCCTCCTCCCGGGCCTCCGCGTTATTTGTTACGCGGACGACACGTTGGTCGTGGCCCGGGGGGATGATTTTAGGGAGTCTGCCCGTCTCGCCACAGCGGGAGTGGCCCTCGTCGTCGGAAGGATAAGGAGGCTGGGTCTCGACGTGGCGCTCAATAAATCCGAGGCTCTGTGGGGTCGGGGTGCAGTTGAAGTACCTCGGCCTCGTGTTGGACAGCCGGTGGGCCTTTCGTGCTCACTTTGCGGAGCTGGTCCCCGATTGATGGGGACGGCCGGTTCTTTGAGCCGGCTGCTCCCGAATATTGGGGGACCGGATCAGGTTGTGCGCCGTCTCTACGCGGGGGTGGTGCGATCGATGGCCCTGTACGGTGCACCTGTGTGGGCTGAGCCGCGCAACCGGGCCACTATGGCTCGGTTCCTGCGCCGGCCGCAGCGCACCGTTGCCATCAGGGTCATCCGCGGATATCGCACCGTCTCCTTCGAGGCGGCGTGTGTTTTGGCGGGGACGCCGCCATGGGAGCTGGAGGCGGAGTCGCTCGCTGCCGACTATCGGTGGCGCAGCGAGCTTCGTGCTCGGGGCGTGGCGCGTGTCCCCGAGAGTGAGCTGCGGGCGCGGAAGGCCCATTCTCGGCGGTCCGTGCTCGAGTCGTGGTCGAGGCGATTGGCCAACCCCACGTGGGGGCTACGGACCGTCGAGGCGGTTTACCCGGTCTTTGATGACTGGGTGAATCGTGGCGAGGGACGTCTCACCTTTCGTCTGGTGCAGGTGCTGACCGGGCACGGATGCTTCGGGAAGTACCTGCGCCGGATAGGGGCTGAGCCGACGACGAGGTGTCACCATTGTGGACACGACCTGGACACGGCGGAGCATACGCTCGCTGTCTGCCCCGCTTGGGAGGTGCAGCGCCGTGTCCTGGTCGCAAAGATAGGACCTGACTTGTCGCTGCCTGGCGTCGTGGCGTCGATGCTTGGCAGCGATGAGTCATGGAAGGCTATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGAGTGAGGGAAAGCTCTCCTCACTACGCAGAAACCCGCCGCCGCCGAGCAGGGGGTCGGGACCGGGGTCGTATCCGTGACCTGGCCCCCTAA
Protein
MVTRGPGIVAVQLENIVVIGVYFSPNRPTVEFERFLDGLEVIARHLAPRSVILAGDFNAKSVAWGSPFTDARGRLLEEWAVAVGLCIVNRGSIATCVRWTGESIVDLTFASSPVARRILGWRVVVGAESLSDHRYIRYDLSAPAPTPAAGARGVDPPQSASRSFPRWALKLLNRELLVEASMVAAWAPKRPQPVDVETEAEWFRGAMHRICDASMPRVGSRHSGRRQSPWWSPEIARLRAVSIRASRRCTRHRRRRRLRRDEPVAFAEEEARLHREYRAAKDALRLAIRRAKEQNMERLLEALEADPWGSPYKLVRGKLRPWAPSMTERLQPQQLRDIVSALFPQEREGFVPPAMGGPSDTVDGQAPAEVPRITEAELRVAVVKMTAKDTAPGPDGVHGRVLALALGALGDRLLELYNGCLESGRFPSLWRTGRLVLLRKEGRPVDTAAGYRPIVLLDEAGKLLERILAARIVRHLVGVGPDLSAEQYGFREGRSTVDAILRVRSLSDEAVSRGGVALAVSLDIANAFNTLPWTVIGGALERHGVPLYLRRLVGSYLGARSVVCTGYGGTLHRFPVVRGVPQGSVLGPLLWNIGYDWVLRGALLPGLRVICYADDTLVVARGDDFRESARLATAGVALVVGRIRRLGLDVALNKSEALWGRGAVEVPRPRVGQPVGLSCSLCGAGPRLMGTAGSLSRLLPNIGGPDQVVRRLYAGVVRSMALYGAPVWAEPRNRATMARFLRRPQRTVAIRVIRGYRTVSFEAACVLAGTPPWELEAESLAADYRWRSELRARGVARVPESELRARKAHSRRSVLESWSRRLANPTWGLRTVEAVYPVFDDWVNRGEGRLTFRLVQVLTGHGCFGKYLRRIGAEPTTRCHHCGHDLDTAEHTLAVCPAWEVQRRVLVAKIGPDLSLPGVVASMLGSDESWKAMLDFCECTISQKEAAGRVRESSPHYAETRRRRAGGRDRGRIRDLAP
Summary
Uniprot
Q868Q4
Q8MY31
Q8MY33
Q8MY35
Q8MY27
A0A2W1C3J5
+ More
A0A0J7N1D9 O01419 Q8MY25 A0A0J7KJG6 A0A0J7KIY5 A0A0J7KQK3 A0A3S2LNH8 A0A0J7KF37 A0A0J7KIM6 A0A0J7KJH4 A0A0J7K3F1 A0A0J7KPR0 A0A0J7N0R3 A0A0J7KYD2 A0A0J7KFH2 A0A0J7MY17 A0A0J7KRB8 A0A0J7KF40 A0A0J7NFZ0 A0A2S2PEZ7 A0A0J7KQY4 A0A0J7KIS5 A0A2S2NQQ6 A0A0J7NAV8 A0A0J7MWN5 A0A0J7KWU5 X1WJY4 A0A2H8TIE5 A0A0J7KMG5 J9KNN8 F7IYV5 A0A2S2PRL8 J9KJG8 A0A0J7KF91 J9LVU7 X1WZ64 J9KM84 X1WXT5 A0A0J7K3R6 A0A2S2P7D1 A0A0J7KPZ3 A0A0J7KLH8 F7IYU8 A0A2S2NJ82 J9LJ80 J9KNS1 A0A139WA31 J9KUM6 J9KT61 A0A2M4BC28 D6WC19 W8ANK7 W8ADE4 J9LM37 F7IYV7 J9JY94 T1DCM0 A0A139W940 A0A0J7KLT9 A0A142LX43 A0A139W9B3 A0A2S2N8I2 J9K6F0 A0A0K8VPH1 A0A2M4BBV9 A0A2M4BC01 A0A0J7KCE1 J9LA43 T1DG59 A0A0J7N8R4 A0A2M4BC18 A0A139W939 A0A2M4BDI3 T1DG89 J9KWM9 F7IYV2 A0A2M4BB89 A0A2M4CS98 A0A2M4BB78 A0A2M4BC24 A0A2M4ADW9 A0A2M4CSA9
A0A0J7N1D9 O01419 Q8MY25 A0A0J7KJG6 A0A0J7KIY5 A0A0J7KQK3 A0A3S2LNH8 A0A0J7KF37 A0A0J7KIM6 A0A0J7KJH4 A0A0J7K3F1 A0A0J7KPR0 A0A0J7N0R3 A0A0J7KYD2 A0A0J7KFH2 A0A0J7MY17 A0A0J7KRB8 A0A0J7KF40 A0A0J7NFZ0 A0A2S2PEZ7 A0A0J7KQY4 A0A0J7KIS5 A0A2S2NQQ6 A0A0J7NAV8 A0A0J7MWN5 A0A0J7KWU5 X1WJY4 A0A2H8TIE5 A0A0J7KMG5 J9KNN8 F7IYV5 A0A2S2PRL8 J9KJG8 A0A0J7KF91 J9LVU7 X1WZ64 J9KM84 X1WXT5 A0A0J7K3R6 A0A2S2P7D1 A0A0J7KPZ3 A0A0J7KLH8 F7IYU8 A0A2S2NJ82 J9LJ80 J9KNS1 A0A139WA31 J9KUM6 J9KT61 A0A2M4BC28 D6WC19 W8ANK7 W8ADE4 J9LM37 F7IYV7 J9JY94 T1DCM0 A0A139W940 A0A0J7KLT9 A0A142LX43 A0A139W9B3 A0A2S2N8I2 J9K6F0 A0A0K8VPH1 A0A2M4BBV9 A0A2M4BC01 A0A0J7KCE1 J9LA43 T1DG59 A0A0J7N8R4 A0A2M4BC18 A0A139W939 A0A2M4BDI3 T1DG89 J9KWM9 F7IYV2 A0A2M4BB89 A0A2M4CS98 A0A2M4BB78 A0A2M4BC24 A0A2M4ADW9 A0A2M4CSA9
EMBL
AB090825
BAC57926.1
AB078931
BAC06456.1
AB078930
BAC06454.1
+ More
AB078929 BAC06452.1 AB078935 BAC06462.1 KZ149896 PZC78733.1 LBMM01012027 KMQ86470.1 D85594 BAA19776.1 AB078936 BAC06464.1 LBMM01006671 KMQ90407.1 LBMM01006954 KMQ90166.1 LBMM01004272 KMQ92618.1 RSAL01003504 RVE40273.1 LBMM01008448 KMQ88867.1 LBMM01007036 KMQ90099.1 LBMM01006657 KMQ90417.1 LBMM01015025 KMQ84953.1 LBMM01004634 KMQ92224.1 LBMM01012412 KMQ86235.1 LBMM01002052 KMQ95336.1 LBMM01008278 KMQ89009.1 LBMM01014122 KMQ85315.1 LBMM01003988 KMQ92922.1 LBMM01008422 KMQ88894.1 LBMM01005474 KMQ91485.1 GGMR01015159 MBY27778.1 LBMM01004262 KMQ92649.1 LBMM01006977 KMQ90149.1 GGMR01006838 MBY19457.1 LBMM01007425 KMQ89740.1 LBMM01015285 KMQ84870.1 LBMM01002480 KMQ94776.1 ABLF02011238 GFXV01002090 MBW13895.1 LBMM01005465 KMQ91492.1 ABLF02023257 AB593325 BAK38646.1 GGMR01019493 MBY32112.1 ABLF02041886 LBMM01008445 KMQ88871.1 ABLF02034925 ABLF02011183 ABLF02041884 ABLF02018942 ABLF02041885 ABLF02004854 ABLF02041317 LBMM01015018 KMQ84954.1 GGMR01012748 MBY25367.1 LBMM01004505 KMQ92346.1 LBMM01005948 KMQ91066.1 AB593321 BAK38639.1 GGMR01004600 MBY17219.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 ABLF02018808 KQ971675 KYB24771.1 ABLF02041557 ABLF02009073 ABLF02030702 ABLF02042963 GGFJ01001439 MBW50580.1 KQ971309 EEZ99146.1 GAMC01020517 JAB86038.1 GAMC01020519 GAMC01020515 JAB86040.1 ABLF02014650 AB593327 BAK38648.1 ABLF02007989 GALA01001777 JAA93075.1 KQ972691 KXZ75806.1 LBMM01005733 KMQ91252.1 KU543682 AMS38369.1 KQ972544 KXZ75869.1 GGMR01000779 MBY13398.1 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1 GGFJ01001369 MBW50510.1 GGFJ01001370 MBW50511.1 LBMM01009535 KMQ88053.1 ABLF02018098 GALA01000302 JAA94550.1 LBMM01008226 KMQ89040.1 GGFJ01001453 MBW50594.1 KXZ75807.1 GGFJ01001727 MBW50868.1 GALA01001776 JAA93076.1 ABLF02011927 AB593323 BAK38643.1 GGFJ01001165 MBW50306.1 GGFL01004044 MBW68222.1 GGFJ01001164 MBW50305.1 GGFJ01001454 MBW50595.1 GGFK01005660 MBW38981.1 GGFL01004046 MBW68224.1
AB078929 BAC06452.1 AB078935 BAC06462.1 KZ149896 PZC78733.1 LBMM01012027 KMQ86470.1 D85594 BAA19776.1 AB078936 BAC06464.1 LBMM01006671 KMQ90407.1 LBMM01006954 KMQ90166.1 LBMM01004272 KMQ92618.1 RSAL01003504 RVE40273.1 LBMM01008448 KMQ88867.1 LBMM01007036 KMQ90099.1 LBMM01006657 KMQ90417.1 LBMM01015025 KMQ84953.1 LBMM01004634 KMQ92224.1 LBMM01012412 KMQ86235.1 LBMM01002052 KMQ95336.1 LBMM01008278 KMQ89009.1 LBMM01014122 KMQ85315.1 LBMM01003988 KMQ92922.1 LBMM01008422 KMQ88894.1 LBMM01005474 KMQ91485.1 GGMR01015159 MBY27778.1 LBMM01004262 KMQ92649.1 LBMM01006977 KMQ90149.1 GGMR01006838 MBY19457.1 LBMM01007425 KMQ89740.1 LBMM01015285 KMQ84870.1 LBMM01002480 KMQ94776.1 ABLF02011238 GFXV01002090 MBW13895.1 LBMM01005465 KMQ91492.1 ABLF02023257 AB593325 BAK38646.1 GGMR01019493 MBY32112.1 ABLF02041886 LBMM01008445 KMQ88871.1 ABLF02034925 ABLF02011183 ABLF02041884 ABLF02018942 ABLF02041885 ABLF02004854 ABLF02041317 LBMM01015018 KMQ84954.1 GGMR01012748 MBY25367.1 LBMM01004505 KMQ92346.1 LBMM01005948 KMQ91066.1 AB593321 BAK38639.1 GGMR01004600 MBY17219.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 ABLF02018808 KQ971675 KYB24771.1 ABLF02041557 ABLF02009073 ABLF02030702 ABLF02042963 GGFJ01001439 MBW50580.1 KQ971309 EEZ99146.1 GAMC01020517 JAB86038.1 GAMC01020519 GAMC01020515 JAB86040.1 ABLF02014650 AB593327 BAK38648.1 ABLF02007989 GALA01001777 JAA93075.1 KQ972691 KXZ75806.1 LBMM01005733 KMQ91252.1 KU543682 AMS38369.1 KQ972544 KXZ75869.1 GGMR01000779 MBY13398.1 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1 GGFJ01001369 MBW50510.1 GGFJ01001370 MBW50511.1 LBMM01009535 KMQ88053.1 ABLF02018098 GALA01000302 JAA94550.1 LBMM01008226 KMQ89040.1 GGFJ01001453 MBW50594.1 KXZ75807.1 GGFJ01001727 MBW50868.1 GALA01001776 JAA93076.1 ABLF02011927 AB593323 BAK38643.1 GGFJ01001165 MBW50306.1 GGFL01004044 MBW68222.1 GGFJ01001164 MBW50305.1 GGFJ01001454 MBW50595.1 GGFK01005660 MBW38981.1 GGFL01004046 MBW68224.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
Q868Q4
Q8MY31
Q8MY33
Q8MY35
Q8MY27
A0A2W1C3J5
+ More
A0A0J7N1D9 O01419 Q8MY25 A0A0J7KJG6 A0A0J7KIY5 A0A0J7KQK3 A0A3S2LNH8 A0A0J7KF37 A0A0J7KIM6 A0A0J7KJH4 A0A0J7K3F1 A0A0J7KPR0 A0A0J7N0R3 A0A0J7KYD2 A0A0J7KFH2 A0A0J7MY17 A0A0J7KRB8 A0A0J7KF40 A0A0J7NFZ0 A0A2S2PEZ7 A0A0J7KQY4 A0A0J7KIS5 A0A2S2NQQ6 A0A0J7NAV8 A0A0J7MWN5 A0A0J7KWU5 X1WJY4 A0A2H8TIE5 A0A0J7KMG5 J9KNN8 F7IYV5 A0A2S2PRL8 J9KJG8 A0A0J7KF91 J9LVU7 X1WZ64 J9KM84 X1WXT5 A0A0J7K3R6 A0A2S2P7D1 A0A0J7KPZ3 A0A0J7KLH8 F7IYU8 A0A2S2NJ82 J9LJ80 J9KNS1 A0A139WA31 J9KUM6 J9KT61 A0A2M4BC28 D6WC19 W8ANK7 W8ADE4 J9LM37 F7IYV7 J9JY94 T1DCM0 A0A139W940 A0A0J7KLT9 A0A142LX43 A0A139W9B3 A0A2S2N8I2 J9K6F0 A0A0K8VPH1 A0A2M4BBV9 A0A2M4BC01 A0A0J7KCE1 J9LA43 T1DG59 A0A0J7N8R4 A0A2M4BC18 A0A139W939 A0A2M4BDI3 T1DG89 J9KWM9 F7IYV2 A0A2M4BB89 A0A2M4CS98 A0A2M4BB78 A0A2M4BC24 A0A2M4ADW9 A0A2M4CSA9
A0A0J7N1D9 O01419 Q8MY25 A0A0J7KJG6 A0A0J7KIY5 A0A0J7KQK3 A0A3S2LNH8 A0A0J7KF37 A0A0J7KIM6 A0A0J7KJH4 A0A0J7K3F1 A0A0J7KPR0 A0A0J7N0R3 A0A0J7KYD2 A0A0J7KFH2 A0A0J7MY17 A0A0J7KRB8 A0A0J7KF40 A0A0J7NFZ0 A0A2S2PEZ7 A0A0J7KQY4 A0A0J7KIS5 A0A2S2NQQ6 A0A0J7NAV8 A0A0J7MWN5 A0A0J7KWU5 X1WJY4 A0A2H8TIE5 A0A0J7KMG5 J9KNN8 F7IYV5 A0A2S2PRL8 J9KJG8 A0A0J7KF91 J9LVU7 X1WZ64 J9KM84 X1WXT5 A0A0J7K3R6 A0A2S2P7D1 A0A0J7KPZ3 A0A0J7KLH8 F7IYU8 A0A2S2NJ82 J9LJ80 J9KNS1 A0A139WA31 J9KUM6 J9KT61 A0A2M4BC28 D6WC19 W8ANK7 W8ADE4 J9LM37 F7IYV7 J9JY94 T1DCM0 A0A139W940 A0A0J7KLT9 A0A142LX43 A0A139W9B3 A0A2S2N8I2 J9K6F0 A0A0K8VPH1 A0A2M4BBV9 A0A2M4BC01 A0A0J7KCE1 J9LA43 T1DG59 A0A0J7N8R4 A0A2M4BC18 A0A139W939 A0A2M4BDI3 T1DG89 J9KWM9 F7IYV2 A0A2M4BB89 A0A2M4CS98 A0A2M4BB78 A0A2M4BC24 A0A2M4ADW9 A0A2M4CSA9
PDB
1WDU
E-value=4.13787e-11,
Score=168
Ontologies
GO
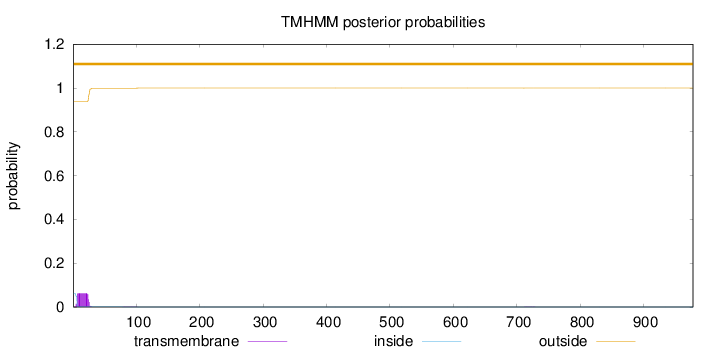
Topology
Length:
978
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.23836000000001
Exp number, first 60 AAs:
1.21235
Total prob of N-in:
0.06282
outside
1 - 978
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0.028447
CSRT
0
Interpretation
Uncertain
