Gene
KWMTBOMO13186
Annotation
PREDICTED:_putative_ATPase_N2B_[Papilio_polytes]
Full name
Putative ATPase N2B
Alternative Name
HFN2B
Location in the cell
Nuclear Reliability : 2.395
Sequence
CDS
ATGTCATACAAAACGACGAAGCTCTGCAATAAGTGTATGAGATTATTATCATCTCAGACTCACGCACAGCACTTCGTAAACGATGGACCTTGGCAAGCATACACTCAGAAAGTGAGCAGCAAGGCATTAAGCAAAGATCCTCATCAAGAACGTGTAGTGCAACATCTGCAAAAAGTATATCAAGAGATTAGTAATTATGAGAGGCCCATAATACAAGAACAAAACATAGGATCGTTTTTTAATTTCTTCAAGAAGCCAGACCCAATAAAGATTGATGCTCCCAAAGGTGTTTACATCTGGGGTAGTGTTGGGGGTGGTAAAACAATGTTAATGGATTTGTTTTATGATACTGTGCCAATTAAAGAAAAGCTCAGAGTTCATTTTAATTCATTTATGTTAAACATCCATGCAAGAATTCATGAGTTAAAAATTAAATCTGGAAAGGGGGCCAGTTCGTTCCGTGATGAAGGTTCTAAACCATTTGATCCTATACCTCCTGTTGCTGCTGATATAACACAGGAATCTTGGCTCATTTGCTTTGATGAATTCCAAGTGACTGATATCGGAGATGCAATGATTCTAAAACGTTTGTTTACTCAGCTCTTTGACAATGGGTGTGTTGTAGTAGCTACAAGCAATAGAAAGCCAGATGATTTGTATAAAAATGGTCTACAAAGATCAAATTTCTTGCCATTCATACCAGTTTTAAAAACACATTGTGATGTTCTACAGTTGGATTCTGGTATTGATTATAGACTACGTGGAAGTGGTACCAAGTACAGTAAATATTTTTTGAATAGTCAAATATCACCTCAAAATGATCCAGTCAATAATTTATTTAAGTTTCTAGTATCAAAGGAGACTGACACTGTGCGGCCTAGAATTATTAATATCTTTGGTAGAAATGTTAAGTTTGCTAAATCCTGTGGAGGGGTTCTTGATTCTACTTTTGAAGAGTTGTGCGATAGACCACTCGGTGCTAGCGATTATTTAGTGATATCAAAAACCTTTCATACTGTGTTCATTAGGAATCTACCTCAATTATCTATTTCGAAGCATAGATCGCAATTGAGAAGATTCATTACACTAATTGATACTTTGTATGATAACAGAGTGAGGGTTGTAATTGCTGCTGATAGCGAACCAAAAAATCTCATGAAATTAGATGAAACTGAATTTGGTGATGCAGACAGAGCACTTATGGATGACTTGAAAATTACTAAAGACTCTGAAGATGCCAAAGCTACTATATTTACGGGTGAAGAAGAAATGTTTGCATGTGATCGATGTCTTTCACGTATAATGGAAATGCAAACCGACGAATATTGGGAGAAATGGGGAACACATATAAACTAA
Protein
MSYKTTKLCNKCMRLLSSQTHAQHFVNDGPWQAYTQKVSSKALSKDPHQERVVQHLQKVYQEISNYERPIIQEQNIGSFFNFFKKPDPIKIDAPKGVYIWGSVGGGKTMLMDLFYDTVPIKEKLRVHFNSFMLNIHARIHELKIKSGKGASSFRDEGSKPFDPIPPVAADITQESWLICFDEFQVTDIGDAMILKRLFTQLFDNGCVVVATSNRKPDDLYKNGLQRSNFLPFIPVLKTHCDVLQLDSGIDYRLRGSGTKYSKYFLNSQISPQNDPVNNLFKFLVSKETDTVRPRIINIFGRNVKFAKSCGGVLDSTFEELCDRPLGASDYLVISKTFHTVFIRNLPQLSISKHRSQLRRFITLIDTLYDNRVRVVIAADSEPKNLMKLDETEFGDADRALMDDLKITKDSEDAKATIFTGEEEMFACDRCLSRIMEMQTDEYWEKWGTHIN
Summary
Similarity
Belongs to the AFG1 ATPase family.
Keywords
ATP-binding
Nucleotide-binding
Feature
chain Putative ATPase N2B
Uniprot
A0A3S2P529
A0A194PX92
A0A194R0L7
A0A212EVA2
H9ISY6
A0A151ILL9
+ More
A0A195F167 A0A158NG09 A0A151XB77 A0A1D2NI69 F4X6I4 E2AHX4 E2B4Z0 A0A182TWI5 A0A2M4AR46 A0A2M4BMG6 A0A2M4AQY8 A0A182PFI7 A0A2M3ZE66 A0A182KTP7 A0A182HRL3 A0A3L8DIG2 W5JAV2 A0A182V550 A0A182F699 A0A182G2I9 A0A182RHF7 A0A182LUF6 A0A195B337 B0WRC6 A0A182K720 A0A0N8ES18 A0A182IRL0 A0A195E7Z4 A0A182Q508 A0A182NUK1 A0A182YDM1 A0A182SJF5 A0A026WT54 Q7Q1B6 A0A0A9WA85 A0A023F838 A0A224XPN6 A0A1Q3FI63 A0A0K8TNA0 A0A0L7RIR0 A0A232EMF6 A0A067R656 R4G3W2 A0A0P4VXY7 A0A336L0S1 T1J356 D6WCA1 A0A088AHT7 U5EXM8 A0A1Y1NFR1 A0A2P8XG13 A0A154PU58 A0A2A3EA34 A0A0M9A8A8 A0A084W218 W8CE98 A0A2R7VQ40 A0A1L8DAM5 A0A1B0GJ93 A0A0C9R4J1 A0A182X786 A0A336K5M8 A0A0J7KGX9 A0A1B6CUE1 A0A1I8MR24 A0A1B6CQJ3 A0A0A1WEY1 T1PEC0 P46441 A0A034W8U2 A0A0K8VYD7 B3MCK5 A0A1I8NX47 B4MQL5 A0A0L0CAQ1 A0A0M4EB94 A7SR07 A0A2L2Y9Y5 A0A182WGT8 E0VAC4 U4UIP3 A0A0P5ZT04 A0A0P5PGZ0 B4GD03 Q292P4 E9HPQ9 A0A0P6J0I6 A0A1A9WJ01 A0A0P5TJA9 A0A0P5UY80 V5HSJ7 A0A0P6B7T4 B4J743
A0A195F167 A0A158NG09 A0A151XB77 A0A1D2NI69 F4X6I4 E2AHX4 E2B4Z0 A0A182TWI5 A0A2M4AR46 A0A2M4BMG6 A0A2M4AQY8 A0A182PFI7 A0A2M3ZE66 A0A182KTP7 A0A182HRL3 A0A3L8DIG2 W5JAV2 A0A182V550 A0A182F699 A0A182G2I9 A0A182RHF7 A0A182LUF6 A0A195B337 B0WRC6 A0A182K720 A0A0N8ES18 A0A182IRL0 A0A195E7Z4 A0A182Q508 A0A182NUK1 A0A182YDM1 A0A182SJF5 A0A026WT54 Q7Q1B6 A0A0A9WA85 A0A023F838 A0A224XPN6 A0A1Q3FI63 A0A0K8TNA0 A0A0L7RIR0 A0A232EMF6 A0A067R656 R4G3W2 A0A0P4VXY7 A0A336L0S1 T1J356 D6WCA1 A0A088AHT7 U5EXM8 A0A1Y1NFR1 A0A2P8XG13 A0A154PU58 A0A2A3EA34 A0A0M9A8A8 A0A084W218 W8CE98 A0A2R7VQ40 A0A1L8DAM5 A0A1B0GJ93 A0A0C9R4J1 A0A182X786 A0A336K5M8 A0A0J7KGX9 A0A1B6CUE1 A0A1I8MR24 A0A1B6CQJ3 A0A0A1WEY1 T1PEC0 P46441 A0A034W8U2 A0A0K8VYD7 B3MCK5 A0A1I8NX47 B4MQL5 A0A0L0CAQ1 A0A0M4EB94 A7SR07 A0A2L2Y9Y5 A0A182WGT8 E0VAC4 U4UIP3 A0A0P5ZT04 A0A0P5PGZ0 B4GD03 Q292P4 E9HPQ9 A0A0P6J0I6 A0A1A9WJ01 A0A0P5TJA9 A0A0P5UY80 V5HSJ7 A0A0P6B7T4 B4J743
Pubmed
26354079
22118469
19121390
21347285
27289101
21719571
+ More
20798317 20966253 30249741 20920257 23761445 26483478 26999592 25244985 24508170 12364791 25401762 26823975 25474469 26369729 28648823 24845553 27129103 18362917 19820115 28004739 29403074 24438588 24495485 25315136 25830018 8551518 25348373 17994087 26108605 17615350 26561354 20566863 23537049 15632085 21292972 25765539
20798317 20966253 30249741 20920257 23761445 26483478 26999592 25244985 24508170 12364791 25401762 26823975 25474469 26369729 28648823 24845553 27129103 18362917 19820115 28004739 29403074 24438588 24495485 25315136 25830018 8551518 25348373 17994087 26108605 17615350 26561354 20566863 23537049 15632085 21292972 25765539
EMBL
RSAL01000387
RVE42021.1
KQ459586
KPI97942.1
KQ460883
KPJ11338.1
+ More
AGBW02012225 OWR45413.1 BABH01013025 BABH01013026 KQ977104 KYN05785.1 KQ981864 KYN34208.1 ADTU01014874 KQ982335 KYQ57589.1 LJIJ01000032 ODN04963.1 GL888818 EGI57944.1 GL439602 EFN66964.1 GL445685 EFN89198.1 GGFK01009871 MBW43192.1 GGFJ01005104 MBW54245.1 GGFK01009870 MBW43191.1 GGFM01006044 MBW26795.1 APCN01001719 QOIP01000008 RLU19992.1 ADMH02002020 ETN59970.1 JXUM01039798 JXUM01039799 JXUM01039800 JXUM01039801 JXUM01039802 KQ561218 KXJ79316.1 AXCM01008671 KQ976662 KYM78604.1 DS232054 EDS33316.1 GDUN01000706 JAN95213.1 KQ979568 KYN20939.1 AXCN02002063 KK107109 EZA59118.1 AAAB01008980 EAA14419.4 GBHO01038930 GBRD01009808 GDHC01000789 JAG04674.1 JAG56016.1 JAQ17840.1 GBBI01001304 JAC17408.1 GFTR01006303 JAW10123.1 GFDL01007789 JAV27256.1 GDAI01002000 JAI15603.1 KQ414583 KOC70719.1 NNAY01003363 OXU19544.1 KK852716 KDR17842.1 ACPB03017886 GAHY01001870 JAA75640.1 GDKW01002145 JAI54450.1 UFQS01001422 UFQT01001422 SSX10806.1 SSX30487.1 JH431820 KQ971316 EEZ98783.1 GANO01002470 JAB57401.1 GEZM01007741 JAV94956.1 PYGN01002284 PSN30912.1 KQ435119 KZC14740.1 KZ288310 PBC28585.1 KQ435727 KOX77909.1 ATLV01019473 KE525272 KFB44262.1 GAMC01000571 JAC05985.1 KK854024 PTY09642.1 GFDF01010561 JAV03523.1 AJWK01019925 GBYB01002835 GBYB01008755 JAG72602.1 JAG78522.1 UFQS01000134 UFQT01000134 SSX00276.1 SSX20656.1 LBMM01007728 KMQ89476.1 GEDC01020271 GEDC01002658 JAS17027.1 JAS34640.1 GEDC01021492 JAS15806.1 GBXI01017056 JAC97235.1 KA647034 AFP61663.1 U12392 GAKP01008382 JAC50570.1 GDHF01008442 JAI43872.1 CH902619 EDV36239.1 CH963849 EDW74404.1 JRES01000678 KNC29326.1 CP012524 ALC42484.1 DS469753 EDO33877.1 IAAA01015268 LAA04961.1 DS235005 EEB10330.1 KB632375 ERL93889.1 GDIP01039961 LRGB01002140 JAM63754.1 KZS08941.1 GDIQ01138842 JAL12884.1 CH479181 EDW31541.1 CM000071 EAL24818.2 GL732709 EFX66268.1 GDIQ01014627 JAN80110.1 GDIP01131219 JAL72495.1 GDIP01106949 JAL96765.1 GANP01007365 JAB77103.1 GDIP01019139 JAM84576.1 CH916367 EDW01031.1
AGBW02012225 OWR45413.1 BABH01013025 BABH01013026 KQ977104 KYN05785.1 KQ981864 KYN34208.1 ADTU01014874 KQ982335 KYQ57589.1 LJIJ01000032 ODN04963.1 GL888818 EGI57944.1 GL439602 EFN66964.1 GL445685 EFN89198.1 GGFK01009871 MBW43192.1 GGFJ01005104 MBW54245.1 GGFK01009870 MBW43191.1 GGFM01006044 MBW26795.1 APCN01001719 QOIP01000008 RLU19992.1 ADMH02002020 ETN59970.1 JXUM01039798 JXUM01039799 JXUM01039800 JXUM01039801 JXUM01039802 KQ561218 KXJ79316.1 AXCM01008671 KQ976662 KYM78604.1 DS232054 EDS33316.1 GDUN01000706 JAN95213.1 KQ979568 KYN20939.1 AXCN02002063 KK107109 EZA59118.1 AAAB01008980 EAA14419.4 GBHO01038930 GBRD01009808 GDHC01000789 JAG04674.1 JAG56016.1 JAQ17840.1 GBBI01001304 JAC17408.1 GFTR01006303 JAW10123.1 GFDL01007789 JAV27256.1 GDAI01002000 JAI15603.1 KQ414583 KOC70719.1 NNAY01003363 OXU19544.1 KK852716 KDR17842.1 ACPB03017886 GAHY01001870 JAA75640.1 GDKW01002145 JAI54450.1 UFQS01001422 UFQT01001422 SSX10806.1 SSX30487.1 JH431820 KQ971316 EEZ98783.1 GANO01002470 JAB57401.1 GEZM01007741 JAV94956.1 PYGN01002284 PSN30912.1 KQ435119 KZC14740.1 KZ288310 PBC28585.1 KQ435727 KOX77909.1 ATLV01019473 KE525272 KFB44262.1 GAMC01000571 JAC05985.1 KK854024 PTY09642.1 GFDF01010561 JAV03523.1 AJWK01019925 GBYB01002835 GBYB01008755 JAG72602.1 JAG78522.1 UFQS01000134 UFQT01000134 SSX00276.1 SSX20656.1 LBMM01007728 KMQ89476.1 GEDC01020271 GEDC01002658 JAS17027.1 JAS34640.1 GEDC01021492 JAS15806.1 GBXI01017056 JAC97235.1 KA647034 AFP61663.1 U12392 GAKP01008382 JAC50570.1 GDHF01008442 JAI43872.1 CH902619 EDV36239.1 CH963849 EDW74404.1 JRES01000678 KNC29326.1 CP012524 ALC42484.1 DS469753 EDO33877.1 IAAA01015268 LAA04961.1 DS235005 EEB10330.1 KB632375 ERL93889.1 GDIP01039961 LRGB01002140 JAM63754.1 KZS08941.1 GDIQ01138842 JAL12884.1 CH479181 EDW31541.1 CM000071 EAL24818.2 GL732709 EFX66268.1 GDIQ01014627 JAN80110.1 GDIP01131219 JAL72495.1 GDIP01106949 JAL96765.1 GANP01007365 JAB77103.1 GDIP01019139 JAM84576.1 CH916367 EDW01031.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000007151
UP000005204
UP000078542
+ More
UP000078541 UP000005205 UP000075809 UP000094527 UP000007755 UP000000311 UP000008237 UP000075902 UP000075885 UP000075882 UP000075840 UP000279307 UP000000673 UP000075903 UP000069272 UP000069940 UP000249989 UP000075900 UP000075883 UP000078540 UP000002320 UP000075881 UP000075880 UP000078492 UP000075886 UP000075884 UP000076408 UP000075901 UP000053097 UP000007062 UP000053825 UP000215335 UP000027135 UP000015103 UP000007266 UP000005203 UP000245037 UP000076502 UP000242457 UP000053105 UP000030765 UP000092461 UP000076407 UP000036403 UP000095301 UP000007801 UP000095300 UP000007798 UP000037069 UP000092553 UP000001593 UP000075920 UP000009046 UP000030742 UP000076858 UP000008744 UP000001819 UP000000305 UP000091820 UP000001070
UP000078541 UP000005205 UP000075809 UP000094527 UP000007755 UP000000311 UP000008237 UP000075902 UP000075885 UP000075882 UP000075840 UP000279307 UP000000673 UP000075903 UP000069272 UP000069940 UP000249989 UP000075900 UP000075883 UP000078540 UP000002320 UP000075881 UP000075880 UP000078492 UP000075886 UP000075884 UP000076408 UP000075901 UP000053097 UP000007062 UP000053825 UP000215335 UP000027135 UP000015103 UP000007266 UP000005203 UP000245037 UP000076502 UP000242457 UP000053105 UP000030765 UP000092461 UP000076407 UP000036403 UP000095301 UP000007801 UP000095300 UP000007798 UP000037069 UP000092553 UP000001593 UP000075920 UP000009046 UP000030742 UP000076858 UP000008744 UP000001819 UP000000305 UP000091820 UP000001070
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A3S2P529
A0A194PX92
A0A194R0L7
A0A212EVA2
H9ISY6
A0A151ILL9
+ More
A0A195F167 A0A158NG09 A0A151XB77 A0A1D2NI69 F4X6I4 E2AHX4 E2B4Z0 A0A182TWI5 A0A2M4AR46 A0A2M4BMG6 A0A2M4AQY8 A0A182PFI7 A0A2M3ZE66 A0A182KTP7 A0A182HRL3 A0A3L8DIG2 W5JAV2 A0A182V550 A0A182F699 A0A182G2I9 A0A182RHF7 A0A182LUF6 A0A195B337 B0WRC6 A0A182K720 A0A0N8ES18 A0A182IRL0 A0A195E7Z4 A0A182Q508 A0A182NUK1 A0A182YDM1 A0A182SJF5 A0A026WT54 Q7Q1B6 A0A0A9WA85 A0A023F838 A0A224XPN6 A0A1Q3FI63 A0A0K8TNA0 A0A0L7RIR0 A0A232EMF6 A0A067R656 R4G3W2 A0A0P4VXY7 A0A336L0S1 T1J356 D6WCA1 A0A088AHT7 U5EXM8 A0A1Y1NFR1 A0A2P8XG13 A0A154PU58 A0A2A3EA34 A0A0M9A8A8 A0A084W218 W8CE98 A0A2R7VQ40 A0A1L8DAM5 A0A1B0GJ93 A0A0C9R4J1 A0A182X786 A0A336K5M8 A0A0J7KGX9 A0A1B6CUE1 A0A1I8MR24 A0A1B6CQJ3 A0A0A1WEY1 T1PEC0 P46441 A0A034W8U2 A0A0K8VYD7 B3MCK5 A0A1I8NX47 B4MQL5 A0A0L0CAQ1 A0A0M4EB94 A7SR07 A0A2L2Y9Y5 A0A182WGT8 E0VAC4 U4UIP3 A0A0P5ZT04 A0A0P5PGZ0 B4GD03 Q292P4 E9HPQ9 A0A0P6J0I6 A0A1A9WJ01 A0A0P5TJA9 A0A0P5UY80 V5HSJ7 A0A0P6B7T4 B4J743
A0A195F167 A0A158NG09 A0A151XB77 A0A1D2NI69 F4X6I4 E2AHX4 E2B4Z0 A0A182TWI5 A0A2M4AR46 A0A2M4BMG6 A0A2M4AQY8 A0A182PFI7 A0A2M3ZE66 A0A182KTP7 A0A182HRL3 A0A3L8DIG2 W5JAV2 A0A182V550 A0A182F699 A0A182G2I9 A0A182RHF7 A0A182LUF6 A0A195B337 B0WRC6 A0A182K720 A0A0N8ES18 A0A182IRL0 A0A195E7Z4 A0A182Q508 A0A182NUK1 A0A182YDM1 A0A182SJF5 A0A026WT54 Q7Q1B6 A0A0A9WA85 A0A023F838 A0A224XPN6 A0A1Q3FI63 A0A0K8TNA0 A0A0L7RIR0 A0A232EMF6 A0A067R656 R4G3W2 A0A0P4VXY7 A0A336L0S1 T1J356 D6WCA1 A0A088AHT7 U5EXM8 A0A1Y1NFR1 A0A2P8XG13 A0A154PU58 A0A2A3EA34 A0A0M9A8A8 A0A084W218 W8CE98 A0A2R7VQ40 A0A1L8DAM5 A0A1B0GJ93 A0A0C9R4J1 A0A182X786 A0A336K5M8 A0A0J7KGX9 A0A1B6CUE1 A0A1I8MR24 A0A1B6CQJ3 A0A0A1WEY1 T1PEC0 P46441 A0A034W8U2 A0A0K8VYD7 B3MCK5 A0A1I8NX47 B4MQL5 A0A0L0CAQ1 A0A0M4EB94 A7SR07 A0A2L2Y9Y5 A0A182WGT8 E0VAC4 U4UIP3 A0A0P5ZT04 A0A0P5PGZ0 B4GD03 Q292P4 E9HPQ9 A0A0P6J0I6 A0A1A9WJ01 A0A0P5TJA9 A0A0P5UY80 V5HSJ7 A0A0P6B7T4 B4J743
Ontologies
GO
PANTHER
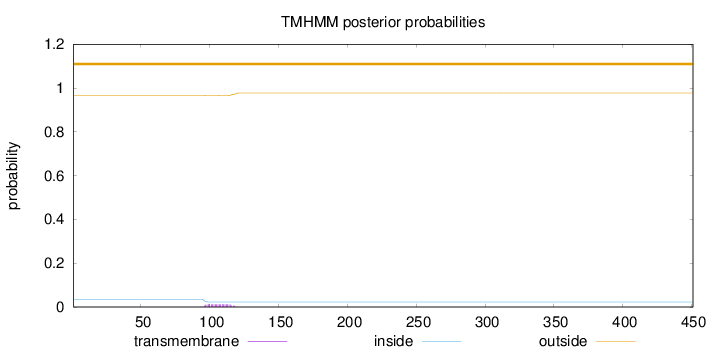
Topology
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.251460000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03459
outside
1 - 451
Population Genetic Test Statistics
Pi
121.618022
Theta
133.664717
Tajima's D
-0.417464
CLR
214.74699
CSRT
0.263536823158842
Interpretation
Uncertain
