Gene
KWMTBOMO13185
Pre Gene Modal
BGIBMGA000355
Annotation
PREDICTED:_general_transcription_factor_IIE_subunit_1_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.319
Sequence
CDS
ATGACGGAAGAGCGTTACGTGACTGAAGTTCCTAGTAGTTTAAAACAGCTAGCAAGATTAGTCGTCAGAGGCTTCTACACAATAGAGGATGCACTCATTGTTGATATGCTGGTCAGGAATCCATGTATGAAAGAAGATGATATTTGTGAGTTATTGAAATTTGAAAGAAAAATGCTAAGAGCAAGAATTTCGATTCTCAAAAATGACAAATTCATTCAGGTCAGACTGAAAATGGAGACAGGACTAGACGGAAAAGCACAAAAAGTTAATTATTATTTCATAAACTACAAGACTTTCGTAAACGTCGTGAAATATAAATTAGATCTTATGAGGAAGCGGTTAGAAACAGAGGAGCGCGACGCTACAAGCAGAGCCAGTTTCAAGTGTCCAGCTTGTGGGAAGACCTTTACAGATTTAGAAGCAGATCAGTTATACGATATGGCAACACAAGAATTCCAATGCACGTTCTGCAGTGCAGTTGTGGACGAGGACATGTCGGCGCTACCAAAAAAAGACTCCAGATTATTGTTGGCCAAGTTCAATGAGCAGCTGGAAACGCTTTACATATTGTTGCGAGAGGTCGAAGGCATCAAACTGGCTCCAGAGATACTGGAACCAGAACCGGTCGATATTAACACAATACGAGGATTGACGTCAAAGCAGTCTGCATTGCGTCCGGGCGGCGAGCAATGGTCGGGTGAGGCGACGCGCAACCAGGGCATGCTGGTCGAGGAGACACGAGTCGACGTCACCATCGGAGACGACAAGCCCGCGCGGGACGCCGGCGCGCTACGCAAGGAGCGGCCCGTCTGGATGGTCGAGAGCACCATTGCCTCCAACGAACAGAGCGACAGCGCGCACTCGACGGACGCGGCCCTCGAGAAGGCGGCCAGTACTGCTGCGAGCGTCAAGACCGCAGGGAAGGAGAAGAACGACGACATTATGTCCGTCCTGCTCGCACACGAGAAACAACCGAGCGCAGGAAACCCTGTCTCAAACGCTGTTAAGGGTCTCGAAGCAGAGAGCTCGGACTCCAGCGACAACGAATCGAAAGATCCGTACAAACTCAAAGATGAACTTGCAGCTGTCGCTGAAATGGAAAGCGATGATTCGGATTCTGATGATAACGCGCCCTCGGTTATGGTGAACGGTAAGACGGTGCCATTGACCAGTGTCGACGACGACGTTATCGCACAGATGACGCCCACTGAGAAGGAAACGTACATCCAAATATATCAGGAATATTACAGCCACATGTACGATTAG
Protein
MTEERYVTEVPSSLKQLARLVVRGFYTIEDALIVDMLVRNPCMKEDDICELLKFERKMLRARISILKNDKFIQVRLKMETGLDGKAQKVNYYFINYKTFVNVVKYKLDLMRKRLETEERDATSRASFKCPACGKTFTDLEADQLYDMATQEFQCTFCSAVVDEDMSALPKKDSRLLLAKFNEQLETLYILLREVEGIKLAPEILEPEPVDINTIRGLTSKQSALRPGGEQWSGEATRNQGMLVEETRVDVTIGDDKPARDAGALRKERPVWMVESTIASNEQSDSAHSTDAALEKAASTAASVKTAGKEKNDDIMSVLLAHEKQPSAGNPVSNAVKGLEAESSDSSDNESKDPYKLKDELAAVAEMESDDSDSDDNAPSVMVNGKTVPLTSVDDDVIAQMTPTEKETYIQIYQEYYSHMYD
Summary
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
H9ISX7
A0A3S2N5F5
S4PX37
A0A194R1E4
A0A194PX63
A0A1E1WTZ6
+ More
A0A212EV90 A0A2P8XFX5 A0A067R446 A0A2J7RJG8 A0A195E7X5 A0A0L7RIC2 E2AHX6 A0A0J7L5E1 A0A026WSV4 A0A154PS60 A0A151ILM8 F4X6I2 A0A195F0V6 A0A088AHJ8 A0A2A3EAT4 E2BAG8 A0A0M9A5Y7 A0A232FAQ2 A0A158NG07 K7IUT8 A0A0C9RTJ8 A0A182QFA2 A0A1Q3F3P8 A0A1W4X097 E9J600 A0A182IWW3 A0A182KCC6 N6UBP6 Q16MT1 U4U9V1 A0A182NK98 A0A182YR26 B0XL91 A0A182PQ94 W5JJE3 A0A182FH78 A0A1B6H7P4 A0A182HI07 A0A182ULP3 A0A182TRI2 A0A182LL95 A0A182WTR1 Q5TR07 A0A182WCC9 A0A182MMI2 D6WNN0 A0A084WEK2 B4J2J0 E0VIK4 A0A1B6KQ19 B4N4M3 B3M4A6 T1IC26 Q2LZ72 A0A1W4W7E9 B4LCQ5 A0A1B6E3X9 B4PFM4 B3NGY8 A0A023F9G6 A0A3B0J506 A0A151XB62 A0A1Y1K3V1 A0A0K8TW87 B4HEW9 B4QQ94 A0A1B0CWW6 A0A0M4F183 A0A1L8DH09 A0A2J7RJH0 A0A069DTZ9 O96880 A0A0P4VKM9 B4KUS0 A0A0A1XSZ6 A0A1B6G783 A0A182GXH3 R4FRA4 A0A182RI94 A0A0L0CE92 W8ARR2 A0A0A9Y1I7 A0A1A9URP5 A0A1B0A6Y4 A0A224XPY1 A0A1B0GG71 A0A336M2K2 A0A336LQN2 A0A1A9XB47 A0A1B0B8T4 A0A1I8PU33 A0A1I8MGX1
A0A212EV90 A0A2P8XFX5 A0A067R446 A0A2J7RJG8 A0A195E7X5 A0A0L7RIC2 E2AHX6 A0A0J7L5E1 A0A026WSV4 A0A154PS60 A0A151ILM8 F4X6I2 A0A195F0V6 A0A088AHJ8 A0A2A3EAT4 E2BAG8 A0A0M9A5Y7 A0A232FAQ2 A0A158NG07 K7IUT8 A0A0C9RTJ8 A0A182QFA2 A0A1Q3F3P8 A0A1W4X097 E9J600 A0A182IWW3 A0A182KCC6 N6UBP6 Q16MT1 U4U9V1 A0A182NK98 A0A182YR26 B0XL91 A0A182PQ94 W5JJE3 A0A182FH78 A0A1B6H7P4 A0A182HI07 A0A182ULP3 A0A182TRI2 A0A182LL95 A0A182WTR1 Q5TR07 A0A182WCC9 A0A182MMI2 D6WNN0 A0A084WEK2 B4J2J0 E0VIK4 A0A1B6KQ19 B4N4M3 B3M4A6 T1IC26 Q2LZ72 A0A1W4W7E9 B4LCQ5 A0A1B6E3X9 B4PFM4 B3NGY8 A0A023F9G6 A0A3B0J506 A0A151XB62 A0A1Y1K3V1 A0A0K8TW87 B4HEW9 B4QQ94 A0A1B0CWW6 A0A0M4F183 A0A1L8DH09 A0A2J7RJH0 A0A069DTZ9 O96880 A0A0P4VKM9 B4KUS0 A0A0A1XSZ6 A0A1B6G783 A0A182GXH3 R4FRA4 A0A182RI94 A0A0L0CE92 W8ARR2 A0A0A9Y1I7 A0A1A9URP5 A0A1B0A6Y4 A0A224XPY1 A0A1B0GG71 A0A336M2K2 A0A336LQN2 A0A1A9XB47 A0A1B0B8T4 A0A1I8PU33 A0A1I8MGX1
Pubmed
19121390
23622113
26354079
22118469
29403074
24845553
+ More
20798317 24508170 30249741 21719571 28648823 21347285 20075255 21282665 23537049 17510324 25244985 20920257 23761445 20966253 12364791 14747013 17210077 18362917 19820115 24438588 17994087 20566863 15632085 17550304 25474469 28004739 22936249 26334808 9012800 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 25830018 26483478 26108605 24495485 25401762 26823975 25315136
20798317 24508170 30249741 21719571 28648823 21347285 20075255 21282665 23537049 17510324 25244985 20920257 23761445 20966253 12364791 14747013 17210077 18362917 19820115 24438588 17994087 20566863 15632085 17550304 25474469 28004739 22936249 26334808 9012800 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 25830018 26483478 26108605 24495485 25401762 26823975 25315136
EMBL
BABH01013026
RSAL01000387
RVE42022.1
GAIX01005718
JAA86842.1
KQ460883
+ More
KPJ11339.1 KQ459586 KPI97941.1 GDQN01000535 JAT90519.1 AGBW02012225 OWR45412.1 PYGN01002284 PSN30913.1 KK852716 KDR17841.1 NEVH01002992 PNF40978.1 KQ979568 KYN20937.1 KQ414583 KOC70717.1 GL439602 EFN66966.1 LBMM01000666 KMQ97841.1 KK107109 QOIP01000008 EZA59120.1 RLU19849.1 KQ435119 KZC14742.1 KQ977104 KYN05787.1 GL888818 EGI57942.1 KQ981864 KYN34210.1 KZ288310 PBC28584.1 GL446721 EFN87330.1 KQ435727 KOX77907.1 NNAY01000550 OXU27735.1 ADTU01014874 AAZX01002635 GBYB01010846 JAG80613.1 AXCN02000423 GFDL01012865 JAV22180.1 GL768221 EFZ11691.1 APGK01041378 KB740993 ENN76067.1 CH477850 EAT35649.1 KB632256 ERL90714.1 DS234361 EDS33725.1 ADMH02000991 ETN64492.1 GECU01036959 GECU01029636 JAS70747.1 JAS78070.1 APCN01005860 AAAB01008960 EAL40056.3 AXCM01004658 KQ971342 EFA03759.1 ATLV01023220 KE525341 KFB48646.1 CH916366 EDV97075.1 DS235200 EEB13210.1 GEBQ01026429 JAT13548.1 CH964095 EDW79097.1 CH902618 EDV39376.1 ACPB03017886 CH379069 EAL29636.1 CH940647 EDW70947.1 GEDC01004668 GEDC01002118 JAS32630.1 JAS35180.1 CM000159 EDW94173.1 CH954178 EDV51445.1 GBBI01000687 JAC18025.1 OUUW01000002 SPP76777.1 KQ982335 KYQ57587.1 GEZM01099042 JAV53537.1 GDHF01033979 GDHF01031666 JAI18335.1 JAI20648.1 CH480815 EDW41138.1 CM000363 CM002912 EDX10112.1 KMY99038.1 AJWK01032930 AJWK01032931 AJWK01032932 AJWK01032933 CP012525 ALC45050.1 GFDF01008346 JAV05738.1 PNF40976.1 GBGD01001643 JAC87246.1 U72892 AE014296 AY119499 AAD00187.1 AAF50029.1 AAM50153.1 GDKW01000941 JAI55654.1 CH933809 EDW19326.1 GBXI01000126 JAD14166.1 GECZ01011477 JAS58292.1 JXUM01095289 JXUM01095290 KQ564186 KXJ72607.1 GAHY01000014 JAA77496.1 JRES01000502 KNC30570.1 GAMC01015050 JAB91505.1 GBHO01016687 GBRD01008864 GDHC01018239 JAG26917.1 JAG56957.1 JAQ00390.1 GFTR01005916 JAW10510.1 CCAG010023484 UFQT01000448 SSX24442.1 UFQT01000118 SSX20384.1 JXJN01010055
KPJ11339.1 KQ459586 KPI97941.1 GDQN01000535 JAT90519.1 AGBW02012225 OWR45412.1 PYGN01002284 PSN30913.1 KK852716 KDR17841.1 NEVH01002992 PNF40978.1 KQ979568 KYN20937.1 KQ414583 KOC70717.1 GL439602 EFN66966.1 LBMM01000666 KMQ97841.1 KK107109 QOIP01000008 EZA59120.1 RLU19849.1 KQ435119 KZC14742.1 KQ977104 KYN05787.1 GL888818 EGI57942.1 KQ981864 KYN34210.1 KZ288310 PBC28584.1 GL446721 EFN87330.1 KQ435727 KOX77907.1 NNAY01000550 OXU27735.1 ADTU01014874 AAZX01002635 GBYB01010846 JAG80613.1 AXCN02000423 GFDL01012865 JAV22180.1 GL768221 EFZ11691.1 APGK01041378 KB740993 ENN76067.1 CH477850 EAT35649.1 KB632256 ERL90714.1 DS234361 EDS33725.1 ADMH02000991 ETN64492.1 GECU01036959 GECU01029636 JAS70747.1 JAS78070.1 APCN01005860 AAAB01008960 EAL40056.3 AXCM01004658 KQ971342 EFA03759.1 ATLV01023220 KE525341 KFB48646.1 CH916366 EDV97075.1 DS235200 EEB13210.1 GEBQ01026429 JAT13548.1 CH964095 EDW79097.1 CH902618 EDV39376.1 ACPB03017886 CH379069 EAL29636.1 CH940647 EDW70947.1 GEDC01004668 GEDC01002118 JAS32630.1 JAS35180.1 CM000159 EDW94173.1 CH954178 EDV51445.1 GBBI01000687 JAC18025.1 OUUW01000002 SPP76777.1 KQ982335 KYQ57587.1 GEZM01099042 JAV53537.1 GDHF01033979 GDHF01031666 JAI18335.1 JAI20648.1 CH480815 EDW41138.1 CM000363 CM002912 EDX10112.1 KMY99038.1 AJWK01032930 AJWK01032931 AJWK01032932 AJWK01032933 CP012525 ALC45050.1 GFDF01008346 JAV05738.1 PNF40976.1 GBGD01001643 JAC87246.1 U72892 AE014296 AY119499 AAD00187.1 AAF50029.1 AAM50153.1 GDKW01000941 JAI55654.1 CH933809 EDW19326.1 GBXI01000126 JAD14166.1 GECZ01011477 JAS58292.1 JXUM01095289 JXUM01095290 KQ564186 KXJ72607.1 GAHY01000014 JAA77496.1 JRES01000502 KNC30570.1 GAMC01015050 JAB91505.1 GBHO01016687 GBRD01008864 GDHC01018239 JAG26917.1 JAG56957.1 JAQ00390.1 GFTR01005916 JAW10510.1 CCAG010023484 UFQT01000448 SSX24442.1 UFQT01000118 SSX20384.1 JXJN01010055
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000245037
+ More
UP000027135 UP000235965 UP000078492 UP000053825 UP000000311 UP000036403 UP000053097 UP000279307 UP000076502 UP000078542 UP000007755 UP000078541 UP000005203 UP000242457 UP000008237 UP000053105 UP000215335 UP000005205 UP000002358 UP000075886 UP000192223 UP000075880 UP000075881 UP000019118 UP000008820 UP000030742 UP000075884 UP000076408 UP000002320 UP000075885 UP000000673 UP000069272 UP000075840 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075920 UP000075883 UP000007266 UP000030765 UP000001070 UP000009046 UP000007798 UP000007801 UP000015103 UP000001819 UP000192221 UP000008792 UP000002282 UP000008711 UP000268350 UP000075809 UP000001292 UP000000304 UP000092461 UP000092553 UP000000803 UP000009192 UP000069940 UP000249989 UP000075900 UP000037069 UP000078200 UP000092445 UP000092444 UP000092443 UP000092460 UP000095300 UP000095301
UP000027135 UP000235965 UP000078492 UP000053825 UP000000311 UP000036403 UP000053097 UP000279307 UP000076502 UP000078542 UP000007755 UP000078541 UP000005203 UP000242457 UP000008237 UP000053105 UP000215335 UP000005205 UP000002358 UP000075886 UP000192223 UP000075880 UP000075881 UP000019118 UP000008820 UP000030742 UP000075884 UP000076408 UP000002320 UP000075885 UP000000673 UP000069272 UP000075840 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075920 UP000075883 UP000007266 UP000030765 UP000001070 UP000009046 UP000007798 UP000007801 UP000015103 UP000001819 UP000192221 UP000008792 UP000002282 UP000008711 UP000268350 UP000075809 UP000001292 UP000000304 UP000092461 UP000092553 UP000000803 UP000009192 UP000069940 UP000249989 UP000075900 UP000037069 UP000078200 UP000092445 UP000092444 UP000092443 UP000092460 UP000095300 UP000095301
Pfam
Interpro
IPR021600
TFIIE_asu_C
+ More
IPR002853 TFIIE_asu
IPR024550 TFIIEa/SarR/Rpc3_HTH_dom
IPR013083 Znf_RING/FYVE/PHD
IPR017919 TFIIE/TFIIEa_HTH
IPR039997 TFE
IPR013137 Znf_TFIIB
IPR018056 Kringle_CS
IPR020067 Frizzled_dom
IPR017441 Protein_kinase_ATP_BS
IPR013806 Kringle-like
IPR000001 Kringle
IPR011009 Kinase-like_dom_sf
IPR038178 Kringle_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR020635 Tyr_kinase_cat_dom
IPR036790 Frizzled_dom_sf
IPR008266 Tyr_kinase_AS
IPR036265 HIT-like_sf
IPR011146 HIT-like
IPR001310 Histidine_triad_HIT
IPR018800 PRCC
IPR019808 Histidine_triad_CS
IPR002853 TFIIE_asu
IPR024550 TFIIEa/SarR/Rpc3_HTH_dom
IPR013083 Znf_RING/FYVE/PHD
IPR017919 TFIIE/TFIIEa_HTH
IPR039997 TFE
IPR013137 Znf_TFIIB
IPR018056 Kringle_CS
IPR020067 Frizzled_dom
IPR017441 Protein_kinase_ATP_BS
IPR013806 Kringle-like
IPR000001 Kringle
IPR011009 Kinase-like_dom_sf
IPR038178 Kringle_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR020635 Tyr_kinase_cat_dom
IPR036790 Frizzled_dom_sf
IPR008266 Tyr_kinase_AS
IPR036265 HIT-like_sf
IPR011146 HIT-like
IPR001310 Histidine_triad_HIT
IPR018800 PRCC
IPR019808 Histidine_triad_CS
Gene 3D
CDD
ProteinModelPortal
H9ISX7
A0A3S2N5F5
S4PX37
A0A194R1E4
A0A194PX63
A0A1E1WTZ6
+ More
A0A212EV90 A0A2P8XFX5 A0A067R446 A0A2J7RJG8 A0A195E7X5 A0A0L7RIC2 E2AHX6 A0A0J7L5E1 A0A026WSV4 A0A154PS60 A0A151ILM8 F4X6I2 A0A195F0V6 A0A088AHJ8 A0A2A3EAT4 E2BAG8 A0A0M9A5Y7 A0A232FAQ2 A0A158NG07 K7IUT8 A0A0C9RTJ8 A0A182QFA2 A0A1Q3F3P8 A0A1W4X097 E9J600 A0A182IWW3 A0A182KCC6 N6UBP6 Q16MT1 U4U9V1 A0A182NK98 A0A182YR26 B0XL91 A0A182PQ94 W5JJE3 A0A182FH78 A0A1B6H7P4 A0A182HI07 A0A182ULP3 A0A182TRI2 A0A182LL95 A0A182WTR1 Q5TR07 A0A182WCC9 A0A182MMI2 D6WNN0 A0A084WEK2 B4J2J0 E0VIK4 A0A1B6KQ19 B4N4M3 B3M4A6 T1IC26 Q2LZ72 A0A1W4W7E9 B4LCQ5 A0A1B6E3X9 B4PFM4 B3NGY8 A0A023F9G6 A0A3B0J506 A0A151XB62 A0A1Y1K3V1 A0A0K8TW87 B4HEW9 B4QQ94 A0A1B0CWW6 A0A0M4F183 A0A1L8DH09 A0A2J7RJH0 A0A069DTZ9 O96880 A0A0P4VKM9 B4KUS0 A0A0A1XSZ6 A0A1B6G783 A0A182GXH3 R4FRA4 A0A182RI94 A0A0L0CE92 W8ARR2 A0A0A9Y1I7 A0A1A9URP5 A0A1B0A6Y4 A0A224XPY1 A0A1B0GG71 A0A336M2K2 A0A336LQN2 A0A1A9XB47 A0A1B0B8T4 A0A1I8PU33 A0A1I8MGX1
A0A212EV90 A0A2P8XFX5 A0A067R446 A0A2J7RJG8 A0A195E7X5 A0A0L7RIC2 E2AHX6 A0A0J7L5E1 A0A026WSV4 A0A154PS60 A0A151ILM8 F4X6I2 A0A195F0V6 A0A088AHJ8 A0A2A3EAT4 E2BAG8 A0A0M9A5Y7 A0A232FAQ2 A0A158NG07 K7IUT8 A0A0C9RTJ8 A0A182QFA2 A0A1Q3F3P8 A0A1W4X097 E9J600 A0A182IWW3 A0A182KCC6 N6UBP6 Q16MT1 U4U9V1 A0A182NK98 A0A182YR26 B0XL91 A0A182PQ94 W5JJE3 A0A182FH78 A0A1B6H7P4 A0A182HI07 A0A182ULP3 A0A182TRI2 A0A182LL95 A0A182WTR1 Q5TR07 A0A182WCC9 A0A182MMI2 D6WNN0 A0A084WEK2 B4J2J0 E0VIK4 A0A1B6KQ19 B4N4M3 B3M4A6 T1IC26 Q2LZ72 A0A1W4W7E9 B4LCQ5 A0A1B6E3X9 B4PFM4 B3NGY8 A0A023F9G6 A0A3B0J506 A0A151XB62 A0A1Y1K3V1 A0A0K8TW87 B4HEW9 B4QQ94 A0A1B0CWW6 A0A0M4F183 A0A1L8DH09 A0A2J7RJH0 A0A069DTZ9 O96880 A0A0P4VKM9 B4KUS0 A0A0A1XSZ6 A0A1B6G783 A0A182GXH3 R4FRA4 A0A182RI94 A0A0L0CE92 W8ARR2 A0A0A9Y1I7 A0A1A9URP5 A0A1B0A6Y4 A0A224XPY1 A0A1B0GG71 A0A336M2K2 A0A336LQN2 A0A1A9XB47 A0A1B0B8T4 A0A1I8PU33 A0A1I8MGX1
PDB
6O9L
E-value=4.01363e-83,
Score=785
Ontologies
GO
PANTHER
Topology
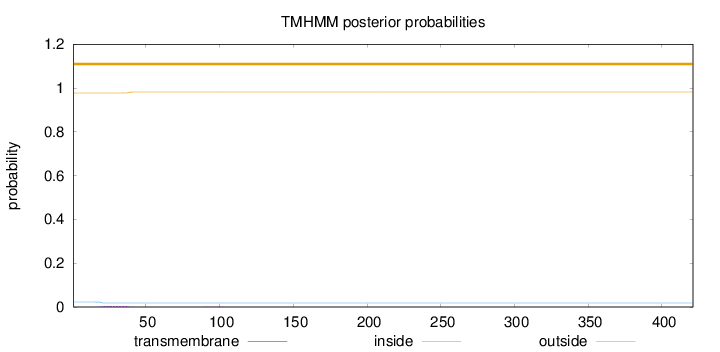
Length:
421
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0887100000000001
Exp number, first 60 AAs:
0.08525
Total prob of N-in:
0.02270
outside
1 - 421
Population Genetic Test Statistics
Pi
207.675317
Theta
163.008804
Tajima's D
0.737776
CLR
0.145193
CSRT
0.586620668966552
Interpretation
Uncertain
