Gene
KWMTBOMO13179 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008821
Annotation
fatty_alcohol_acetyltransferase_[Agrotis_segetum]
Full name
Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex
+ More
Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial
Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial
Alternative Name
Branched-chain alpha-keto acid dehydrogenase complex component E2
Dihydrolipoamide acetyltransferase component of branched-chain alpha-keto acid dehydrogenase complex
Dihydrolipoamide branched chain transacylase
Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase
Dihydrolipoamide acetyltransferase component of branched-chain alpha-keto acid dehydrogenase complex
Dihydrolipoamide branched chain transacylase
Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase
Location in the cell
Mitochondrial Reliability : 2.384
Sequence
CDS
ATGTCGATTCTTGTTCGTAGATCTGTATTCCAACTGAGAACCGTGAACCGGTGCCGGAAAATCAAGGTGACGACGCAAAGTGCGAGAAACGGATCACAGCTATCTTACAAAACGCCTTTGAATGAAAGCTTAAGCAAGGAGTTAAGGCATTTCCACACCAGTCATGCTGTCAACAAAATCGTCGCTTTCAAGCTTTCTGATATCGGTGAAGGGATACGAGAAGTCGTCATCAAGGAATGGTTCGTCAAAGTCGGTGATAATGTTCAACAATTCGACAACATTTGCGAAGTACAAAGCGACAAGGCAGCTGTCACCATCACTAGCCGATACGACGGTATCATAACACGACTGTACCATGATATTGATCAGACCGCTCTTGTAGGACAACCCCTAGTCGATATTGACGTGCAAGACTCTGAAAATGACGGGAAACCCACAGACGTTGCGCCTGATAAGCCTGTTGCTGAAATAGACGCTCCGAAAACGGAACAAAACCAAAAAATTAAAGTATTAACTACTCCGGCGGTGAGAAGAATCGCCGCTCAATTTAAAGTAGACTTGAGTGCCGTTAAAGCAACTGGCAGAAACGGAAGAGTTCTCAAGGAAGACATGTTGTCTCATTTAAACATTAACTCGGATGACTCGAACGAAGTCGTATTACCTGGTAGTGTCAGAGCTGTAGAAATACCAGTTACAAAAGCGCAGGCAAGAGTAGAATTAGCTCTCGAGGATAAAGTTGTCCCTGTTACCGGCTTCCAGAGAGCAATGGTTAAATCTATGACGGAGGCCATGAAAATTCCGCACTTTGTATTCAGTGACGAGTACAACGTCACAAAACTGGTTGAATCGCGTGCAGCACTCAAAAAGCAAGCGGAAGAACGTGGGGTTAGACTAACTTACATGCCGATCATCATCAAAGCGGCATCTTTAAGTTTGAGCTATCTACCCATTCTCAACAGTAGTCTCGATAGCTCCTGTGAAAATCTCATTTACAAAGCAAGTCATAACATAGGCGTTGCTATGGATACGCCCAACGGATTAGTCGTACCTGTTATCAAGAACATCCAAATTAAATCAATATTAGAAATTGCACAAGAGTTAAATACATTGCAAGAGAAAGGATCGAAGGGGCAACTTGGATTGAATGATCTCACCGGAGGGACATTTACAATATCAAATATCGGAATTGTTGGAGGTACTTACACGAAACCGGTGATATTCCCACCGCAAGTTGCAATTGGAGCATTAGGAAAAATACAGGTTAGTAGTGCGTAG
Protein
MSILVRRSVFQLRTVNRCRKIKVTTQSARNGSQLSYKTPLNESLSKELRHFHTSHAVNKIVAFKLSDIGEGIREVVIKEWFVKVGDNVQQFDNICEVQSDKAAVTITSRYDGIITRLYHDIDQTALVGQPLVDIDVQDSENDGKPTDVAPDKPVAEIDAPKTEQNQKIKVLTTPAVRRIAAQFKVDLSAVKATGRNGRVLKEDMLSHLNINSDDSNEVVLPGSVRAVEIPVTKAQARVELALEDKVVPVTGFQRAMVKSMTEAMKIPHFVFSDEYNVTKLVESRAALKKQAEERGVRLTYMPIIIKAASLSLSYLPILNSSLDSSCENLIYKASHNIGVAMDTPNGLVVPVIKNIQIKSILEIAQELNTLQEKGSKGQLGLNDLTGGTFTISNIGIVGGTYTKPVIFPPQVAIGALGKIQVSSA
Summary
Catalytic Activity
(R)-N(6)-dihydrolipoyl-L-lysyl-[protein] + 2-methylpropanoyl-CoA = (R)-N(6)-(S(8)-2-methylpropanoyldihydrolipoyl)-L-lysyl-[protein] + CoA
Cofactor
(R)-lipoate
Subunit
Forms a 24-polypeptide structural core with octahedral symmetry.
Similarity
Belongs to the 2-oxoacid dehydrogenase family.
Keywords
Acetylation
Acyltransferase
Complete proteome
Lipoyl
Mitochondrion
Phosphoprotein
Reference proteome
Transferase
Transit peptide
Feature
chain Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial
Uniprot
H9JH24
A0A291P133
A0A291P181
A0A088ML95
A0A1V0M886
A0A0P0E5H1
+ More
A0A2H1W075 A0A2A4IW66 A0A3S2KZ82 A0A212EV79 A0A1L8D6H5 A0A194R1E9 A0A194PX56 H0Z485 A0A093BWL8 A0A091KHD4 A0A091PIJ9 A0A093F9D0 A0A091QET6 A0A2I0MDV3 A0A091QHG9 A0A094KWW3 A0A091MLK6 A0A3L8SPN8 A0A093QPE4 A0A1V4JYG2 A0A091NI61 A0A091HGW8 G1N527 A0A091WEC9 A0A1V4JYN9 A0A087VGJ2 H0Z400 A0A1L8E0H2 A0A0A0AG34 A0A093I6X5 A0A093NB19 A0A093J441 A0A091VDF2 A0A2I0UID2 A0A099Z4G6 A0A1L8E0E2 A0A218VF26 A0A2D0RWA8 A0A091SJB9 R0JWK3 A0A091HU72 Q98UJ6 A0A091EHH1 A0A0Q3MCQ4 A0A1D5PWD8 A0A093GNZ4 A0A2C9JUM3 A0A091MJ87 A0A093S9S3 A0A3Q1D146 A0A091G9W2 A0A3B4EL58 A0A3P8S0J9 A0A091S7G3 A0A1I7TTQ4 A0A1I7YGF7 A0A1I7TTQ3 A0A1L8GN47 D2HFE5 A0A1B0C967 Q642P5 W5LJ32 A0A3Q0FZU5 A0A182YQ37 A0A151M4T6 A0A151M4T4 W5LW66 A0A1L8GGF6 A0A1W4Y2E9 A0A2D0RD28 G3SY24 A0A3B4D8C3 A0A3B4CZ64 A0A090LHD6 A0A3Q1F9B3 A0A182W4N9 A0A182JSU6 A0A0R4INZ8 W5LFA4 A0A182M3L0 A0A3P4S2X9 A0A3M0K5H9 Q5BKV3 Q7TND9 A0A3B4Y8P4 A0A1L8GGF4 A0A2Y9SB63 A0A2M4BLG5 A0A2Y9SIS1 P53395 E3LG82 Q28E15 A0A182FIU4 A0A182P7W6
A0A2H1W075 A0A2A4IW66 A0A3S2KZ82 A0A212EV79 A0A1L8D6H5 A0A194R1E9 A0A194PX56 H0Z485 A0A093BWL8 A0A091KHD4 A0A091PIJ9 A0A093F9D0 A0A091QET6 A0A2I0MDV3 A0A091QHG9 A0A094KWW3 A0A091MLK6 A0A3L8SPN8 A0A093QPE4 A0A1V4JYG2 A0A091NI61 A0A091HGW8 G1N527 A0A091WEC9 A0A1V4JYN9 A0A087VGJ2 H0Z400 A0A1L8E0H2 A0A0A0AG34 A0A093I6X5 A0A093NB19 A0A093J441 A0A091VDF2 A0A2I0UID2 A0A099Z4G6 A0A1L8E0E2 A0A218VF26 A0A2D0RWA8 A0A091SJB9 R0JWK3 A0A091HU72 Q98UJ6 A0A091EHH1 A0A0Q3MCQ4 A0A1D5PWD8 A0A093GNZ4 A0A2C9JUM3 A0A091MJ87 A0A093S9S3 A0A3Q1D146 A0A091G9W2 A0A3B4EL58 A0A3P8S0J9 A0A091S7G3 A0A1I7TTQ4 A0A1I7YGF7 A0A1I7TTQ3 A0A1L8GN47 D2HFE5 A0A1B0C967 Q642P5 W5LJ32 A0A3Q0FZU5 A0A182YQ37 A0A151M4T6 A0A151M4T4 W5LW66 A0A1L8GGF6 A0A1W4Y2E9 A0A2D0RD28 G3SY24 A0A3B4D8C3 A0A3B4CZ64 A0A090LHD6 A0A3Q1F9B3 A0A182W4N9 A0A182JSU6 A0A0R4INZ8 W5LFA4 A0A182M3L0 A0A3P4S2X9 A0A3M0K5H9 Q5BKV3 Q7TND9 A0A3B4Y8P4 A0A1L8GGF4 A0A2Y9SB63 A0A2M4BLG5 A0A2Y9SIS1 P53395 E3LG82 Q28E15 A0A182FIU4 A0A182P7W6
EC Number
2.3.1.-
2.3.1.168
2.3.1.168
Pubmed
EMBL
BABH01037515
BABH01037516
MF687658
ATJ44584.1
MF706187
ATJ44614.1
+ More
KJ579234 AIN34710.1 KU755497 ARD71207.1 KT261719 ALJ30259.1 ODYU01005529 SOQ46468.1 NWSH01006136 PCG63638.1 RSAL01000387 RVE42026.1 AGBW02012225 OWR45408.1 GEYN01000162 JAV01967.1 KQ460883 KPJ11344.1 KQ459586 KPI97936.1 ABQF01035311 KL230344 KFV05254.1 KK502396 KFP23126.1 KK660240 KFQ07742.1 KK385525 KFV50846.1 KK674900 KFQ08034.1 AKCR02000018 PKK27859.1 KK797889 KFQ26653.1 KL269744 KFZ63633.1 KK829556 KFP75414.1 QUSF01000010 RLW05897.1 KL420106 KFW88235.1 LSYS01005497 OPJ77260.1 KL383079 KFP88697.1 KL536886 KFO94714.1 KK735283 KFR13455.1 OPJ77261.1 KL492575 KFO11734.1 ABQF01035305 ABQF01035306 GFDF01001883 JAV12201.1 KL871737 KGL93141.1 KL206922 KFV86882.1 KL224623 KFW62138.1 KK601813 KFW06652.1 KL410872 KFR00423.1 KZ505741 PKU45814.1 KL889294 KGL76616.1 GFDF01001884 JAV12200.1 MUZQ01000002 OWK64519.1 KK474025 KFQ58423.1 KB743021 EOB01911.1 KL217788 KFO98692.1 AB035085 BAB32667.1 KK718445 KFO56531.1 LMAW01002543 KQK80192.1 AADN05000647 KL216678 KFV71028.1 KK506687 KFP61620.1 KL670581 KFW79589.1 KL447874 KFO78131.1 KK944852 KFQ53522.1 CM004472 OCT85257.1 GL192786 EFB14675.1 AJWK01001964 BC081233 AAH81233.1 AKHW03006584 KYO19532.1 KYO19533.1 CM004473 OCT82929.1 LN609529 CEF67553.1 CR751228 CR936359 AXCM01001653 CYRY02045899 VCX41429.1 QRBI01000117 RMC08445.1 BC090917 BC165614 AAH90917.1 AAI65614.1 BC055890 AAH55890.1 OCT82930.1 GGFJ01004778 MBW53919.1 L42996 AK165959 CH466532 DS268408 EFO86267.1 AAMC01030687 CR848496 CAJ82272.1
KJ579234 AIN34710.1 KU755497 ARD71207.1 KT261719 ALJ30259.1 ODYU01005529 SOQ46468.1 NWSH01006136 PCG63638.1 RSAL01000387 RVE42026.1 AGBW02012225 OWR45408.1 GEYN01000162 JAV01967.1 KQ460883 KPJ11344.1 KQ459586 KPI97936.1 ABQF01035311 KL230344 KFV05254.1 KK502396 KFP23126.1 KK660240 KFQ07742.1 KK385525 KFV50846.1 KK674900 KFQ08034.1 AKCR02000018 PKK27859.1 KK797889 KFQ26653.1 KL269744 KFZ63633.1 KK829556 KFP75414.1 QUSF01000010 RLW05897.1 KL420106 KFW88235.1 LSYS01005497 OPJ77260.1 KL383079 KFP88697.1 KL536886 KFO94714.1 KK735283 KFR13455.1 OPJ77261.1 KL492575 KFO11734.1 ABQF01035305 ABQF01035306 GFDF01001883 JAV12201.1 KL871737 KGL93141.1 KL206922 KFV86882.1 KL224623 KFW62138.1 KK601813 KFW06652.1 KL410872 KFR00423.1 KZ505741 PKU45814.1 KL889294 KGL76616.1 GFDF01001884 JAV12200.1 MUZQ01000002 OWK64519.1 KK474025 KFQ58423.1 KB743021 EOB01911.1 KL217788 KFO98692.1 AB035085 BAB32667.1 KK718445 KFO56531.1 LMAW01002543 KQK80192.1 AADN05000647 KL216678 KFV71028.1 KK506687 KFP61620.1 KL670581 KFW79589.1 KL447874 KFO78131.1 KK944852 KFQ53522.1 CM004472 OCT85257.1 GL192786 EFB14675.1 AJWK01001964 BC081233 AAH81233.1 AKHW03006584 KYO19532.1 KYO19533.1 CM004473 OCT82929.1 LN609529 CEF67553.1 CR751228 CR936359 AXCM01001653 CYRY02045899 VCX41429.1 QRBI01000117 RMC08445.1 BC090917 BC165614 AAH90917.1 AAI65614.1 BC055890 AAH55890.1 OCT82930.1 GGFJ01004778 MBW53919.1 L42996 AK165959 CH466532 DS268408 EFO86267.1 AAMC01030687 CR848496 CAJ82272.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000007754 UP000053119 UP000053872 UP000276834 UP000190648 UP000001645 UP000053605 UP000053858 UP000053584 UP000054081 UP000053283 UP000053641 UP000197619 UP000221080 UP000054308 UP000052976 UP000051836 UP000000539 UP000053875 UP000076420 UP000053258 UP000257160 UP000053760 UP000261440 UP000265080 UP000095282 UP000095287 UP000186698 UP000092461 UP000018467 UP000189705 UP000076408 UP000050525 UP000018468 UP000192224 UP000007646 UP000035682 UP000257200 UP000075920 UP000075881 UP000000437 UP000075883 UP000269221 UP000261360 UP000248484 UP000000589 UP000008281 UP000008143 UP000069272 UP000075885
UP000007754 UP000053119 UP000053872 UP000276834 UP000190648 UP000001645 UP000053605 UP000053858 UP000053584 UP000054081 UP000053283 UP000053641 UP000197619 UP000221080 UP000054308 UP000052976 UP000051836 UP000000539 UP000053875 UP000076420 UP000053258 UP000257160 UP000053760 UP000261440 UP000265080 UP000095282 UP000095287 UP000186698 UP000092461 UP000018467 UP000189705 UP000076408 UP000050525 UP000018468 UP000192224 UP000007646 UP000035682 UP000257200 UP000075920 UP000075881 UP000000437 UP000075883 UP000269221 UP000261360 UP000248484 UP000000589 UP000008281 UP000008143 UP000069272 UP000075885
Interpro
Gene 3D
ProteinModelPortal
H9JH24
A0A291P133
A0A291P181
A0A088ML95
A0A1V0M886
A0A0P0E5H1
+ More
A0A2H1W075 A0A2A4IW66 A0A3S2KZ82 A0A212EV79 A0A1L8D6H5 A0A194R1E9 A0A194PX56 H0Z485 A0A093BWL8 A0A091KHD4 A0A091PIJ9 A0A093F9D0 A0A091QET6 A0A2I0MDV3 A0A091QHG9 A0A094KWW3 A0A091MLK6 A0A3L8SPN8 A0A093QPE4 A0A1V4JYG2 A0A091NI61 A0A091HGW8 G1N527 A0A091WEC9 A0A1V4JYN9 A0A087VGJ2 H0Z400 A0A1L8E0H2 A0A0A0AG34 A0A093I6X5 A0A093NB19 A0A093J441 A0A091VDF2 A0A2I0UID2 A0A099Z4G6 A0A1L8E0E2 A0A218VF26 A0A2D0RWA8 A0A091SJB9 R0JWK3 A0A091HU72 Q98UJ6 A0A091EHH1 A0A0Q3MCQ4 A0A1D5PWD8 A0A093GNZ4 A0A2C9JUM3 A0A091MJ87 A0A093S9S3 A0A3Q1D146 A0A091G9W2 A0A3B4EL58 A0A3P8S0J9 A0A091S7G3 A0A1I7TTQ4 A0A1I7YGF7 A0A1I7TTQ3 A0A1L8GN47 D2HFE5 A0A1B0C967 Q642P5 W5LJ32 A0A3Q0FZU5 A0A182YQ37 A0A151M4T6 A0A151M4T4 W5LW66 A0A1L8GGF6 A0A1W4Y2E9 A0A2D0RD28 G3SY24 A0A3B4D8C3 A0A3B4CZ64 A0A090LHD6 A0A3Q1F9B3 A0A182W4N9 A0A182JSU6 A0A0R4INZ8 W5LFA4 A0A182M3L0 A0A3P4S2X9 A0A3M0K5H9 Q5BKV3 Q7TND9 A0A3B4Y8P4 A0A1L8GGF4 A0A2Y9SB63 A0A2M4BLG5 A0A2Y9SIS1 P53395 E3LG82 Q28E15 A0A182FIU4 A0A182P7W6
A0A2H1W075 A0A2A4IW66 A0A3S2KZ82 A0A212EV79 A0A1L8D6H5 A0A194R1E9 A0A194PX56 H0Z485 A0A093BWL8 A0A091KHD4 A0A091PIJ9 A0A093F9D0 A0A091QET6 A0A2I0MDV3 A0A091QHG9 A0A094KWW3 A0A091MLK6 A0A3L8SPN8 A0A093QPE4 A0A1V4JYG2 A0A091NI61 A0A091HGW8 G1N527 A0A091WEC9 A0A1V4JYN9 A0A087VGJ2 H0Z400 A0A1L8E0H2 A0A0A0AG34 A0A093I6X5 A0A093NB19 A0A093J441 A0A091VDF2 A0A2I0UID2 A0A099Z4G6 A0A1L8E0E2 A0A218VF26 A0A2D0RWA8 A0A091SJB9 R0JWK3 A0A091HU72 Q98UJ6 A0A091EHH1 A0A0Q3MCQ4 A0A1D5PWD8 A0A093GNZ4 A0A2C9JUM3 A0A091MJ87 A0A093S9S3 A0A3Q1D146 A0A091G9W2 A0A3B4EL58 A0A3P8S0J9 A0A091S7G3 A0A1I7TTQ4 A0A1I7YGF7 A0A1I7TTQ3 A0A1L8GN47 D2HFE5 A0A1B0C967 Q642P5 W5LJ32 A0A3Q0FZU5 A0A182YQ37 A0A151M4T6 A0A151M4T4 W5LW66 A0A1L8GGF6 A0A1W4Y2E9 A0A2D0RD28 G3SY24 A0A3B4D8C3 A0A3B4CZ64 A0A090LHD6 A0A3Q1F9B3 A0A182W4N9 A0A182JSU6 A0A0R4INZ8 W5LFA4 A0A182M3L0 A0A3P4S2X9 A0A3M0K5H9 Q5BKV3 Q7TND9 A0A3B4Y8P4 A0A1L8GGF4 A0A2Y9SB63 A0A2M4BLG5 A0A2Y9SIS1 P53395 E3LG82 Q28E15 A0A182FIU4 A0A182P7W6
PDB
2II5
E-value=2.3125e-61,
Score=597
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion matrix
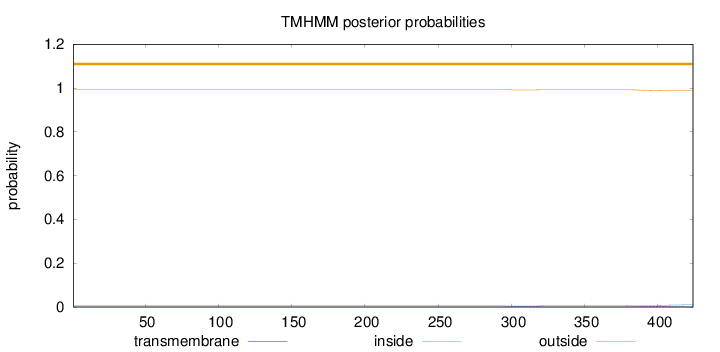
Length:
424
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18913
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00590
outside
1 - 424
Population Genetic Test Statistics
Pi
220.840547
Theta
199.336007
Tajima's D
0.378305
CLR
1.329939
CSRT
0.479926003699815
Interpretation
Uncertain
