Gene
KWMTBOMO13175
Pre Gene Modal
BGIBMGA000366
Annotation
PREDICTED:_probable_cation-transporting_ATPase_13A3_[Bombyx_mori]
Full name
Cation-transporting ATPase
Location in the cell
PlasmaMembrane Reliability : 4.803
Sequence
CDS
ATGCCTTTTTTCAAAAATGATCCTCTCGCCAATGATGAATATGAAGTCATCGGTGAAGCGTTTTGGCAGACCCTGGAAATTCCCGGAGAAGAACAAATTTTAAAAATATATGGCTACAAATATTCGAAGATGCATGCCCTCGTTTTCCACTCTATATGTACAGTATTATGTGGAGTCCCATACATTGCATTGCTGTACTACCCAAAGTACAATCGATTCAAATATGTGAAATGCAGTTTAAAATCAGCAGAAAAGATTTGCGTGACAGATTCAGATGGATTATTTTATTTAGCACAAATAGATGAAATGGATCTCAATCTATCGAATCATAGAGAAAATAAATTAAGATATTTTATTTACCAGCATAATCGTTACATTTGGTTGAGTGATCAAGGTGCCTTTGTTAATGTTCTCGCTCTCAACGAAAAACTAACTATATGTATGTTGATGGAAAATCTAACAGGACTCAACAAGAGACAACAAAATGAATTGCTGAAACTTTATGGTGGAAATAGTGTGGAGGTAAAAGTAATGAGTTACTGGACCATCTTCATCAATGAAGTCTTTAATCCATTCTATTTGTTCCAAGTTTTCTCCATTGTTCTTTGGTCATTGGATGAGTACTATCAATATGCTACTTGTGTGTTCATTTTGTCAACTGCGTCATGTGTAACAGCACTGTGTCAAACAAAACAGATGAGTATCAAAATTCACAAAATGGCTGGTGGTACTAGTGCTTTTACTGTAACGGTTCTGAGACCAACGCGTTCTGGACGTGAAGAATGCGTGGTGAATGCTAGCAGGCTGGTCCCTGGAGATGTATTAGTGATACCATCTGATGGATGTGTAATGCCTTGTGATGCGGTGATTCTGACGGGTTCTTGTATTGTTAACGAGAGTATGCTAACAGGTGAAAGTGTACCAGTTATGAAAGGCCCTCCGTGTTTGTCATCTGAAGTGTATTCTACAGAAGTTCATAAACGACATACACTCTATGCTGGCACTCATGTGATACAAACCAGATTTTATGGTAATAACCAGGTTTTAGCTAAAGTCGTAAGGACCGGATTTTACACAGCTAAAGGAGAGCTTATAAAAAGCATACTGTATCCAAAAGAGGTCAATTTCCAGTTCTACGACGACGCCGTTAGATTCGTAACATTCATGTTCTCCATTGCTGCGCTTGGCATGGCTTACTCCGTCTGGTTATACGTCCTGCGAGGGAGCTCCGTCAGTACGATAATGCTCCGTACATTGGACATTATAACCATAGTGGTACCGCCTGCACTACCCGCGGCCATGACAGCCGGCGTAGTTTACAGCCAACGCCGCTTAAGGAAAAACAAAATATTCTGCGTCTCACCGCCGAGGATCGTGATTTGCGGGAAATTACAAGTCATGTGCTTTGATAAGACTGGTACTTTAACTGAAGACGGCTTAGACTTCTATTCGGTAATTCCTTCAAATGAGGGCGAGAAATTCGGGAAACAAGTGGTGAATGTGACGGGGCTGCCGGCTACGAGTCCTCTACTGCAGGCCCTGGCTTCGTGTCATTCCTTGACCAGCATCCAAGGCGAGCTTAAAGGAGATCCGCTTGATTTGAAAATGTTTGAGTTTACACGATGGGTACTGGAGGAGCCCGGCCCAGAGAACACGCGGTATGACAACCTAACACCGTCTATAGTTAAACCCGTATCGCCAACGGAGAGCACACAGAATCTTGACAATTATGACCCTTACACAATGGAAATTCCGTATGAGATTGGTCTTCTAAGGAGGTTTCACTTTTCTTCATCCCAACAATCTATGGGAGTTATCGCCCGAGTGCTTGGACAACCACAAATGGTGTATTTTGTTAAAGGAGCGCCCGAGAAGATCTCCGGCATGTGCGCCCCGGAAACCCTGCCGGAAAACTTCAGCACAGTACTTAACGAATACACATCGAACGGCTTCCGCGTGATCGGTCTCGCTCACAAGAAGCTTGACCGTAAAATGAAATGGGTCGACGCCCAGAGAATCAAAAGAGATACCCTCGAATGCGATATGGTCTTCCTAGGCTTCCTAGTCATGCAAAATAGCCTGAAAACCGAAACTACCCAGATCATAAGAGAGCTACACATGGCCAACATGAGGCTGGTCATGGTCACAGGAGATAACATCATGACGGCTATGTCAGTGGCCCGCGGATGCTTCATGGTGCAACCGCAACAGAAGATAGTGTTGATAACTGCAGTGCCACAAGCGAACGACGCTCAACCTCCACTCACCATGGAAGTCGTCGGAGAGAGCGGACCACCAAAGTTACCGATGGATTCTTATGTCATAGCTTTGGAAGGGAAGACTTGGCAAGCAATACGAACGTACTATCCAGATTTAATGCCGACTGTATTGAAAAAAGGTATGGTGTTCGGTCGCTTCGGTCCGGACCAGAAGACGCAGCTGATAACCGCGCTGCAGGACGAGGAGCTGACGGTGGGCATGTGCGGGGACGGCGCCAACGACTGCGGCGCCCTCAAAGCCGCGCACGTCGGGATATCGCTCTCCGAAGCCGACGCTTCCGTGGCGGCACCGTTCACTTCTCAAGAGCAAAACATCACATGCGTCAAACTGCTAGTTCTAGAGGGACGTTGTGCACTCAGCACTTCATTCGCAATATTTAAATATATGGCTCTGTATTCGCTGATACAGTTCTTCTCAATTCTTATTCTATACAAGCACCATTCGACCCTGGGCAACAATCAGTTCCTGTACATCGACCTGGTGCTGACGACGCTGCTGGCGCTGTCCCTGGGCCGCGCATGTCCGGGCCCGCGCCTCACGGCCGCGCTGCCGCCCGTGTCGCTGGTGTCGCCCGTCAGCCTGGCGCAGCTGCTGCTGCAGGTGCTCCTCGTGCTCATGGTCCAGATCTCGGCCATCTACGTGCTCCACGGCCAACCTTGGTTCAAATCCATCGAAGGTGGACCGAACATAGAAAATATAATGTGCTGGGACAACACAGTAATATTCATCACGACCTCGTTTCAGTACTTGATCATGGCCTGTGTTTACGCCAACGGCTGGCCGTTCCGAGACCGCTTTTACGCTAACTATTCGATGGTGCTATCGCTGGTGACGCAAACCTCATTCGTGATACTGCTGTTGATGTGCCCCTGGTCGTGGCTCGCGCATTTCATGGACATCAAAATGTTGGACCCCACAGATCCGGACCAAAAAACTTTCCGTACTTATCTGATGTTGATACCTTTCATACATCTGATCTTGGCTTTTACTATAGAAATGACGTTGTCGAATTCAGAAAAATTCTCGAAGCTGTCGAAGTTGATCCGGAGACGGTGCCGGGACAAGGAGGACGCGTCAGTGGAAGGCGCGTGTCCGCTGTGGGCGCCGCCTGCGCACGTGTGCTGA
Protein
MPFFKNDPLANDEYEVIGEAFWQTLEIPGEEQILKIYGYKYSKMHALVFHSICTVLCGVPYIALLYYPKYNRFKYVKCSLKSAEKICVTDSDGLFYLAQIDEMDLNLSNHRENKLRYFIYQHNRYIWLSDQGAFVNVLALNEKLTICMLMENLTGLNKRQQNELLKLYGGNSVEVKVMSYWTIFINEVFNPFYLFQVFSIVLWSLDEYYQYATCVFILSTASCVTALCQTKQMSIKIHKMAGGTSAFTVTVLRPTRSGREECVVNASRLVPGDVLVIPSDGCVMPCDAVILTGSCIVNESMLTGESVPVMKGPPCLSSEVYSTEVHKRHTLYAGTHVIQTRFYGNNQVLAKVVRTGFYTAKGELIKSILYPKEVNFQFYDDAVRFVTFMFSIAALGMAYSVWLYVLRGSSVSTIMLRTLDIITIVVPPALPAAMTAGVVYSQRRLRKNKIFCVSPPRIVICGKLQVMCFDKTGTLTEDGLDFYSVIPSNEGEKFGKQVVNVTGLPATSPLLQALASCHSLTSIQGELKGDPLDLKMFEFTRWVLEEPGPENTRYDNLTPSIVKPVSPTESTQNLDNYDPYTMEIPYEIGLLRRFHFSSSQQSMGVIARVLGQPQMVYFVKGAPEKISGMCAPETLPENFSTVLNEYTSNGFRVIGLAHKKLDRKMKWVDAQRIKRDTLECDMVFLGFLVMQNSLKTETTQIIRELHMANMRLVMVTGDNIMTAMSVARGCFMVQPQQKIVLITAVPQANDAQPPLTMEVVGESGPPKLPMDSYVIALEGKTWQAIRTYYPDLMPTVLKKGMVFGRFGPDQKTQLITALQDEELTVGMCGDGANDCGALKAAHVGISLSEADASVAAPFTSQEQNITCVKLLVLEGRCALSTSFAIFKYMALYSLIQFFSILILYKHHSTLGNNQFLYIDLVLTTLLALSLGRACPGPRLTAALPPVSLVSPVSLAQLLLQVLLVLMVQISAIYVLHGQPWFKSIEGGPNIENIMCWDNTVIFITTSFQYLIMACVYANGWPFRDRFYANYSMVLSLVTQTSFVILLLMCPWSWLAHFMDIKMLDPTDPDQKTFRTYLMLIPFIHLILAFTIEMTLSNSEKFSKLSKLIRRRCRDKEDASVEGACPLWAPPAHVC
Summary
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type V subfamily.
Feature
chain Cation-transporting ATPase
Uniprot
H9ISY8
A0A2H1VKD8
A0A212EV83
A0A194PY31
A0A194R0W8
A0A2J7PBB3
+ More
A0A139WII3 A0A1W4X573 A0A1Y1NJ34 A0A0P4W4W7 A0A1S3HL31 A0A1B6HL55 A0A087U2A5 A0A0P4VJ67 A0A023F0T4 A0A1B6I5F2 A0A2T7PUC4 A0A210QD82 A0A1B6EJF3 A0A224XDK4 A0A1B6M2J0 K1QW82 A0A0A9WMI9 A0A1Y1NNF1 A0A146KML7 A0A131XUC6 A0A0A9WJX6 A0A2H8TMY9 J9JX04 A0A1B6KE17 A0A2S2PVU9 A0A2B4STL1 A0A1W4XFN0 A0A2R2ML53 T1K1X3 A0A1W4XER6 M3XKU8 K9INY1 U3J4Z5 H2V5N9 H2V5P5 H2V5P0 K9IQ02 A0A3Q0GIH9 A0A1S3JY92 A0A1U8D1P3 A0A1U8DC01 A0A2I0M2U1 H3AXA6 A0A3Q0GJ71 A0A3Q0GMR7 A0A1D5PJ96 A0A3Q2TYV0 A0A151MJ69 A0A3P9NHI1 A0A3P9NHQ3 A0A1D5P4F4 H3AXA5 A0A3P9NHS6 A0A132A2F8 A0A2R5L8D8 A0A1S3FFR1 A0A3Q2Y7L2 A0A1S3FE45 A0A3Q2Y913 H3B7S7 A0A131YX45 A0A224YT27 A0A131YCS1 A0A3P8WCY6 A0A3B4H1M6 A0A1D5PH63 A0A1A7WD17 L7M2P7 A0A3B4H3Q8 A0A315UWB7 L7LSE7 A0A3B4H089 A0A3P8N752 A0A3P8N799 E1BG26 L7MHP4 A0A3Q2V1V7 A0A3Q2WEH3 I3J349 A0A210PXR6 A0A3B3XIT5 A0A3B3YTL7 A0A3Q1LSQ6 A0A3Q1M5T0 A0A3Q1LMJ5 A0A091QHG5 A0A0P6JGF9 A0A091J7U4 A0A250YM52 A0A0K8T8V4 A0A2Y9KAQ9 A0A3B5PRK5 A0A1E1XB83 M3ZTQ6
A0A139WII3 A0A1W4X573 A0A1Y1NJ34 A0A0P4W4W7 A0A1S3HL31 A0A1B6HL55 A0A087U2A5 A0A0P4VJ67 A0A023F0T4 A0A1B6I5F2 A0A2T7PUC4 A0A210QD82 A0A1B6EJF3 A0A224XDK4 A0A1B6M2J0 K1QW82 A0A0A9WMI9 A0A1Y1NNF1 A0A146KML7 A0A131XUC6 A0A0A9WJX6 A0A2H8TMY9 J9JX04 A0A1B6KE17 A0A2S2PVU9 A0A2B4STL1 A0A1W4XFN0 A0A2R2ML53 T1K1X3 A0A1W4XER6 M3XKU8 K9INY1 U3J4Z5 H2V5N9 H2V5P5 H2V5P0 K9IQ02 A0A3Q0GIH9 A0A1S3JY92 A0A1U8D1P3 A0A1U8DC01 A0A2I0M2U1 H3AXA6 A0A3Q0GJ71 A0A3Q0GMR7 A0A1D5PJ96 A0A3Q2TYV0 A0A151MJ69 A0A3P9NHI1 A0A3P9NHQ3 A0A1D5P4F4 H3AXA5 A0A3P9NHS6 A0A132A2F8 A0A2R5L8D8 A0A1S3FFR1 A0A3Q2Y7L2 A0A1S3FE45 A0A3Q2Y913 H3B7S7 A0A131YX45 A0A224YT27 A0A131YCS1 A0A3P8WCY6 A0A3B4H1M6 A0A1D5PH63 A0A1A7WD17 L7M2P7 A0A3B4H3Q8 A0A315UWB7 L7LSE7 A0A3B4H089 A0A3P8N752 A0A3P8N799 E1BG26 L7MHP4 A0A3Q2V1V7 A0A3Q2WEH3 I3J349 A0A210PXR6 A0A3B3XIT5 A0A3B3YTL7 A0A3Q1LSQ6 A0A3Q1M5T0 A0A3Q1LMJ5 A0A091QHG5 A0A0P6JGF9 A0A091J7U4 A0A250YM52 A0A0K8T8V4 A0A2Y9KAQ9 A0A3B5PRK5 A0A1E1XB83 M3ZTQ6
EC Number
3.6.3.-
Pubmed
EMBL
BABH01013034
BABH01013035
ODYU01003023
SOQ41266.1
AGBW02012225
OWR45405.1
+ More
KQ459586 KPI97933.1 KQ460883 KPJ11347.1 NEVH01027130 PNF13620.1 KQ971338 KYB27756.1 GEZM01001379 JAV97914.1 GDRN01101414 JAI58382.1 GECU01032284 GECU01001829 JAS75422.1 JAT05878.1 KK117826 KFM71494.1 GDKW01001599 JAI54996.1 GBBI01003605 JAC15107.1 GECU01028811 GECU01025542 GECU01019835 JAS78895.1 JAS82164.1 JAS87871.1 PZQS01000002 PVD36977.1 NEDP02004099 OWF46704.1 GECZ01031716 GECZ01013366 JAS38053.1 JAS56403.1 GFTR01008558 JAW07868.1 GEBQ01009826 JAT30151.1 JH817540 EKC41107.1 GBHO01035891 JAG07713.1 GEZM01001381 JAV97906.1 GDHC01021310 JAP97318.1 GEFM01005864 JAP69932.1 GBHO01035893 GDHC01011867 JAG07711.1 JAQ06762.1 GFXV01003690 MBW15495.1 ABLF02033530 ABLF02033534 ABLF02040091 ABLF02040104 ABLF02040105 ABLF02040112 ABLF02040113 ABLF02053377 ABLF02055581 ABLF02063237 GEBQ01030332 JAT09645.1 GGMS01000310 MBY69513.1 LSMT01000029 PFX31918.1 CAEY01001357 AFYH01072371 AFYH01072372 AFYH01072373 AFYH01072374 AFYH01072375 GABZ01003832 JAA49693.1 ADON01026100 ADON01026101 GABZ01003778 JAA49747.1 AKCR02000044 PKK24002.1 AFYH01105219 AFYH01105220 AFYH01105221 AFYH01105222 AFYH01105223 AFYH01105224 AADN05000197 AKHW03006061 KYO24581.1 JXLN01010091 KPM05094.1 GGLE01001640 MBY05766.1 GEDV01005477 JAP83080.1 GFPF01006595 MAA17741.1 GEDV01012295 JAP76262.1 HADW01002492 SBP03892.1 GACK01006912 JAA58122.1 NHOQ01002824 PWA14538.1 GACK01010043 JAA54991.1 GACK01002345 JAA62689.1 AERX01037043 NEDP02005415 OWF41249.1 KK699335 KFQ26291.1 GEBF01003806 JAN99826.1 KK501618 KFP16712.1 GFFW01000091 JAV44697.1 GBRD01004257 GBRD01004256 JAG61565.1 GFAC01002690 JAT96498.1
KQ459586 KPI97933.1 KQ460883 KPJ11347.1 NEVH01027130 PNF13620.1 KQ971338 KYB27756.1 GEZM01001379 JAV97914.1 GDRN01101414 JAI58382.1 GECU01032284 GECU01001829 JAS75422.1 JAT05878.1 KK117826 KFM71494.1 GDKW01001599 JAI54996.1 GBBI01003605 JAC15107.1 GECU01028811 GECU01025542 GECU01019835 JAS78895.1 JAS82164.1 JAS87871.1 PZQS01000002 PVD36977.1 NEDP02004099 OWF46704.1 GECZ01031716 GECZ01013366 JAS38053.1 JAS56403.1 GFTR01008558 JAW07868.1 GEBQ01009826 JAT30151.1 JH817540 EKC41107.1 GBHO01035891 JAG07713.1 GEZM01001381 JAV97906.1 GDHC01021310 JAP97318.1 GEFM01005864 JAP69932.1 GBHO01035893 GDHC01011867 JAG07711.1 JAQ06762.1 GFXV01003690 MBW15495.1 ABLF02033530 ABLF02033534 ABLF02040091 ABLF02040104 ABLF02040105 ABLF02040112 ABLF02040113 ABLF02053377 ABLF02055581 ABLF02063237 GEBQ01030332 JAT09645.1 GGMS01000310 MBY69513.1 LSMT01000029 PFX31918.1 CAEY01001357 AFYH01072371 AFYH01072372 AFYH01072373 AFYH01072374 AFYH01072375 GABZ01003832 JAA49693.1 ADON01026100 ADON01026101 GABZ01003778 JAA49747.1 AKCR02000044 PKK24002.1 AFYH01105219 AFYH01105220 AFYH01105221 AFYH01105222 AFYH01105223 AFYH01105224 AADN05000197 AKHW03006061 KYO24581.1 JXLN01010091 KPM05094.1 GGLE01001640 MBY05766.1 GEDV01005477 JAP83080.1 GFPF01006595 MAA17741.1 GEDV01012295 JAP76262.1 HADW01002492 SBP03892.1 GACK01006912 JAA58122.1 NHOQ01002824 PWA14538.1 GACK01010043 JAA54991.1 GACK01002345 JAA62689.1 AERX01037043 NEDP02005415 OWF41249.1 KK699335 KFQ26291.1 GEBF01003806 JAN99826.1 KK501618 KFP16712.1 GFFW01000091 JAV44697.1 GBRD01004257 GBRD01004256 JAG61565.1 GFAC01002690 JAT96498.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000235965
UP000007266
+ More
UP000192223 UP000085678 UP000054359 UP000245119 UP000242188 UP000005408 UP000007819 UP000225706 UP000015104 UP000008672 UP000016666 UP000005226 UP000189705 UP000053872 UP000000539 UP000050525 UP000242638 UP000081671 UP000264820 UP000265120 UP000261460 UP000265100 UP000009136 UP000264840 UP000005207 UP000261480 UP000053119 UP000248482 UP000002852
UP000192223 UP000085678 UP000054359 UP000245119 UP000242188 UP000005408 UP000007819 UP000225706 UP000015104 UP000008672 UP000016666 UP000005226 UP000189705 UP000053872 UP000000539 UP000050525 UP000242638 UP000081671 UP000264820 UP000265120 UP000261460 UP000265100 UP000009136 UP000264840 UP000005207 UP000261480 UP000053119 UP000248482 UP000002852
Interpro
IPR004014
ATPase_P-typ_cation-transptr_N
+ More
IPR018303 ATPase_P-typ_P_site
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006544 P-type_TPase_V
IPR023298 ATPase_P-typ_TM_dom_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR001757 P_typ_ATPase
IPR006068 ATPase_P-typ_cation-transptr_C
IPR008673 MAGP
IPR003582 ShKT_dom
IPR018303 ATPase_P-typ_P_site
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006544 P-type_TPase_V
IPR023298 ATPase_P-typ_TM_dom_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR001757 P_typ_ATPase
IPR006068 ATPase_P-typ_cation-transptr_C
IPR008673 MAGP
IPR003582 ShKT_dom
Gene 3D
ProteinModelPortal
H9ISY8
A0A2H1VKD8
A0A212EV83
A0A194PY31
A0A194R0W8
A0A2J7PBB3
+ More
A0A139WII3 A0A1W4X573 A0A1Y1NJ34 A0A0P4W4W7 A0A1S3HL31 A0A1B6HL55 A0A087U2A5 A0A0P4VJ67 A0A023F0T4 A0A1B6I5F2 A0A2T7PUC4 A0A210QD82 A0A1B6EJF3 A0A224XDK4 A0A1B6M2J0 K1QW82 A0A0A9WMI9 A0A1Y1NNF1 A0A146KML7 A0A131XUC6 A0A0A9WJX6 A0A2H8TMY9 J9JX04 A0A1B6KE17 A0A2S2PVU9 A0A2B4STL1 A0A1W4XFN0 A0A2R2ML53 T1K1X3 A0A1W4XER6 M3XKU8 K9INY1 U3J4Z5 H2V5N9 H2V5P5 H2V5P0 K9IQ02 A0A3Q0GIH9 A0A1S3JY92 A0A1U8D1P3 A0A1U8DC01 A0A2I0M2U1 H3AXA6 A0A3Q0GJ71 A0A3Q0GMR7 A0A1D5PJ96 A0A3Q2TYV0 A0A151MJ69 A0A3P9NHI1 A0A3P9NHQ3 A0A1D5P4F4 H3AXA5 A0A3P9NHS6 A0A132A2F8 A0A2R5L8D8 A0A1S3FFR1 A0A3Q2Y7L2 A0A1S3FE45 A0A3Q2Y913 H3B7S7 A0A131YX45 A0A224YT27 A0A131YCS1 A0A3P8WCY6 A0A3B4H1M6 A0A1D5PH63 A0A1A7WD17 L7M2P7 A0A3B4H3Q8 A0A315UWB7 L7LSE7 A0A3B4H089 A0A3P8N752 A0A3P8N799 E1BG26 L7MHP4 A0A3Q2V1V7 A0A3Q2WEH3 I3J349 A0A210PXR6 A0A3B3XIT5 A0A3B3YTL7 A0A3Q1LSQ6 A0A3Q1M5T0 A0A3Q1LMJ5 A0A091QHG5 A0A0P6JGF9 A0A091J7U4 A0A250YM52 A0A0K8T8V4 A0A2Y9KAQ9 A0A3B5PRK5 A0A1E1XB83 M3ZTQ6
A0A139WII3 A0A1W4X573 A0A1Y1NJ34 A0A0P4W4W7 A0A1S3HL31 A0A1B6HL55 A0A087U2A5 A0A0P4VJ67 A0A023F0T4 A0A1B6I5F2 A0A2T7PUC4 A0A210QD82 A0A1B6EJF3 A0A224XDK4 A0A1B6M2J0 K1QW82 A0A0A9WMI9 A0A1Y1NNF1 A0A146KML7 A0A131XUC6 A0A0A9WJX6 A0A2H8TMY9 J9JX04 A0A1B6KE17 A0A2S2PVU9 A0A2B4STL1 A0A1W4XFN0 A0A2R2ML53 T1K1X3 A0A1W4XER6 M3XKU8 K9INY1 U3J4Z5 H2V5N9 H2V5P5 H2V5P0 K9IQ02 A0A3Q0GIH9 A0A1S3JY92 A0A1U8D1P3 A0A1U8DC01 A0A2I0M2U1 H3AXA6 A0A3Q0GJ71 A0A3Q0GMR7 A0A1D5PJ96 A0A3Q2TYV0 A0A151MJ69 A0A3P9NHI1 A0A3P9NHQ3 A0A1D5P4F4 H3AXA5 A0A3P9NHS6 A0A132A2F8 A0A2R5L8D8 A0A1S3FFR1 A0A3Q2Y7L2 A0A1S3FE45 A0A3Q2Y913 H3B7S7 A0A131YX45 A0A224YT27 A0A131YCS1 A0A3P8WCY6 A0A3B4H1M6 A0A1D5PH63 A0A1A7WD17 L7M2P7 A0A3B4H3Q8 A0A315UWB7 L7LSE7 A0A3B4H089 A0A3P8N752 A0A3P8N799 E1BG26 L7MHP4 A0A3Q2V1V7 A0A3Q2WEH3 I3J349 A0A210PXR6 A0A3B3XIT5 A0A3B3YTL7 A0A3Q1LSQ6 A0A3Q1M5T0 A0A3Q1LMJ5 A0A091QHG5 A0A0P6JGF9 A0A091J7U4 A0A250YM52 A0A0K8T8V4 A0A2Y9KAQ9 A0A3B5PRK5 A0A1E1XB83 M3ZTQ6
PDB
5AW9
E-value=6.47203e-21,
Score=253
Ontologies
GO
GO:0005524
GO:0046872
GO:0006812
GO:0016021
GO:0016887
GO:0005623
GO:0006874
GO:1905123
GO:0097734
GO:0043005
GO:1902047
GO:1905165
GO:1990938
GO:0016243
GO:0016241
GO:0030133
GO:0012506
GO:0006882
GO:0080025
GO:0034599
GO:0043025
GO:0005776
GO:0070300
GO:1903543
GO:0005771
GO:1905103
GO:0052548
GO:0006879
GO:0071287
GO:0050714
GO:0010821
GO:0016787
GO:0001527
GO:0043009
GO:0007626
GO:0000166
GO:0005737
GO:0048869
GO:0003676
GO:0016491
GO:0015930
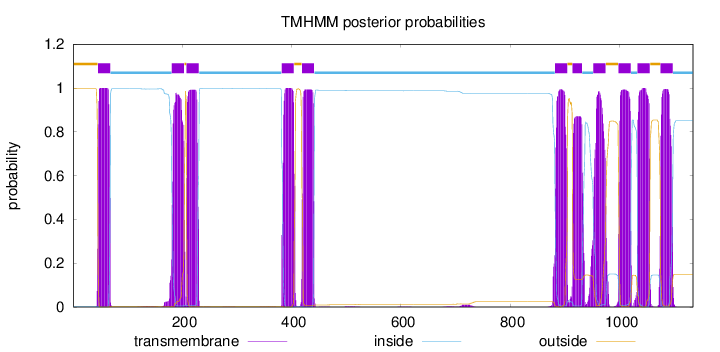
Topology
Subcellular location
Membrane
Length:
1134
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
237.2415
Exp number, first 60 AAs:
15.11079
Total prob of N-in:
0.00298
POSSIBLE N-term signal
sequence
outside
1 - 45
TMhelix
46 - 68
inside
69 - 180
TMhelix
181 - 203
outside
204 - 207
TMhelix
208 - 230
inside
231 - 381
TMhelix
382 - 404
outside
405 - 418
TMhelix
419 - 441
inside
442 - 881
TMhelix
882 - 904
outside
905 - 913
TMhelix
914 - 931
inside
932 - 951
TMhelix
952 - 974
outside
975 - 997
TMhelix
998 - 1020
inside
1021 - 1032
TMhelix
1033 - 1055
outside
1056 - 1074
TMhelix
1075 - 1097
inside
1098 - 1134
Population Genetic Test Statistics
Pi
269.400892
Theta
193.838765
Tajima's D
1.339338
CLR
0.004502
CSRT
0.749362531873406
Interpretation
Uncertain
