Gene
KWMTBOMO13170
Pre Gene Modal
BGIBMGA000350
Annotation
PREDICTED:_uncharacterized_protein_LOC106133968_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.807
Sequence
CDS
ATGCAAGGGCTTGCTAGTTACACAACCCTATCAGTTAGTGTGATGAACCCTCATTTAATGGTCAGGCTCTTCCCGCAAAGAGACATCACGGATGTATTGTTTCTTAGTGCTGGAACTGGGGCAGGTTTATGGCTTTGGGCAAGACCACACATGGCTGCAGCTGTTCCTGCACGTCGATTTTCTTGGGCGTTTTTAGGTGGTTGTCTCTGGCCTTTAGGTTCTATATTTTTATGGGCAATCCTAAGGAGTTCAGTAGGACAAAGGCCAGTCATTGGCACAATAATCGGTGTAACGACTGGAGCTATGCTTACAAATATAGCTTTAGATTATTTCCATTACATAGAACTTATGTTTTGTGGAGAATTACATTTACCTGAAGAAACTTATACCCCGAATAGTGATGATGAAAGATACGATATTGATATTTAA
Protein
MQGLASYTTLSVSVMNPHLMVRLFPQRDITDVLFLSAGTGAGLWLWARPHMAAAVPARRFSWAFLGGCLWPLGSIFLWAILRSSVGQRPVIGTIIGVTTGAMLTNIALDYFHYIELMFCGELHLPEETYTPNSDDERYDIDI
Summary
Uniprot
H9ISX2
A0A1E1VXF9
A0A2H1X3Q6
A0A194PYV6
A0A194R660
S4NNN1
+ More
A0A0L7L6R7 A0A212EV77 A0A2P8XEH7 Q1HQL4 Q16ND9 J3JTX9 A0A0T6BA50 B4GIJ1 D6WH15 Q28YY1 A0A023EF95 A0A1I8Q6D8 A0A1L8EEG4 A0A3B0J177 A0A0K8SHZ8 B4J8H5 A0A0A9WNK3 A0A182FZR7 V5GB57 A0A1W4WSC5 U5EM09 A0A2M4AH27 B0WTC8 A0A1Q3EV05 A0A067RG54 A0A084WHF8 A0A154PHD2 A0A2C9GR40 A0A182ULT8 A0A182TGZ0 A0A182WZX8 A0A182KSY7 A0A0K8U8X5 W8CD06 E9JB68 B8RIX1 A0A0J7KZY8 A0A182P1S3 Q7QKM6 A0A1I8MFE8 A0A2A3E166 V9IKC9 A0A088AA90 T1PCI8 B4MPD4 W5JCB2 A0A182JLB4 A0A310SJU3 A0A034WLK7 E2AVB0 A0A2M3Z7A8 A0A2J7QPV5 A0A182K8H2 A0A0N0BDF9 A0A182T6G9 A0A182MW09 A0A0M4EV79 A0A336MMT1 T1DQE9 A0A0A1XCS5 B4MFM3 A0A0L7QQF9 B4P7H7 A0A1W4UGG9 B4KRQ5 A0A182Y217 A0A0K8TQL5 B4HNE0 B4QB65 Q6NP72 B3NSK0 A0A026WSX4 A0A182W9D7 A0A182N2L5 A0A182QVD3 B3MET9 A0A023EE51 A0A0L0CAU8 E2C7U2 A0A3L8DFM3 A0A1L8D9J2 A0A1B0CRH9 A0A1B6H3Y2 A0A1B6J3K9 A0A1B6DNG9 R4WDB8
A0A0L7L6R7 A0A212EV77 A0A2P8XEH7 Q1HQL4 Q16ND9 J3JTX9 A0A0T6BA50 B4GIJ1 D6WH15 Q28YY1 A0A023EF95 A0A1I8Q6D8 A0A1L8EEG4 A0A3B0J177 A0A0K8SHZ8 B4J8H5 A0A0A9WNK3 A0A182FZR7 V5GB57 A0A1W4WSC5 U5EM09 A0A2M4AH27 B0WTC8 A0A1Q3EV05 A0A067RG54 A0A084WHF8 A0A154PHD2 A0A2C9GR40 A0A182ULT8 A0A182TGZ0 A0A182WZX8 A0A182KSY7 A0A0K8U8X5 W8CD06 E9JB68 B8RIX1 A0A0J7KZY8 A0A182P1S3 Q7QKM6 A0A1I8MFE8 A0A2A3E166 V9IKC9 A0A088AA90 T1PCI8 B4MPD4 W5JCB2 A0A182JLB4 A0A310SJU3 A0A034WLK7 E2AVB0 A0A2M3Z7A8 A0A2J7QPV5 A0A182K8H2 A0A0N0BDF9 A0A182T6G9 A0A182MW09 A0A0M4EV79 A0A336MMT1 T1DQE9 A0A0A1XCS5 B4MFM3 A0A0L7QQF9 B4P7H7 A0A1W4UGG9 B4KRQ5 A0A182Y217 A0A0K8TQL5 B4HNE0 B4QB65 Q6NP72 B3NSK0 A0A026WSX4 A0A182W9D7 A0A182N2L5 A0A182QVD3 B3MET9 A0A023EE51 A0A0L0CAU8 E2C7U2 A0A3L8DFM3 A0A1L8D9J2 A0A1B0CRH9 A0A1B6H3Y2 A0A1B6J3K9 A0A1B6DNG9 R4WDB8
Pubmed
19121390
26354079
23622113
26227816
22118469
29403074
+ More
17204158 17510324 22516182 23537049 17994087 18362917 19820115 15632085 24945155 26483478 26823975 25401762 24845553 24438588 20966253 24495485 21282665 9087549 12364791 14747013 17210077 25315136 20920257 23761445 25348373 20798317 25830018 17550304 25244985 26369729 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 26108605 30249741 23691247
17204158 17510324 22516182 23537049 17994087 18362917 19820115 15632085 24945155 26483478 26823975 25401762 24845553 24438588 20966253 24495485 21282665 9087549 12364791 14747013 17210077 25315136 20920257 23761445 25348373 20798317 25830018 17550304 25244985 26369729 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 26108605 30249741 23691247
EMBL
BABH01013035
GDQN01011693
JAT79361.1
ODYU01013257
SOQ59961.1
KQ459586
+ More
KPI97929.1 KQ460883 KPJ11351.1 GAIX01013861 JAA78699.1 JTDY01002647 KOB71011.1 AGBW02012225 OWR45400.1 PYGN01002489 PSN30399.1 DQ440430 ABF18463.1 CH477828 EAT35860.1 APGK01057485 BT126689 KB741280 KB632033 AEE61651.1 ENN70944.1 ERL88184.1 LJIG01009094 KRT83767.1 CH479183 EDW36311.1 KQ971321 EFA00618.1 CM000071 EAL25834.1 JXUM01095302 GAPW01005525 GAPW01005524 KQ564187 JAC08073.1 KXJ72604.1 GFDG01001697 JAV17102.1 OUUW01000001 SPP74595.1 GBRD01013421 GBRD01013420 GBRD01013419 GDHC01020311 GDHC01014732 JAG52405.1 JAP98317.1 JAQ03897.1 CH916367 EDW02334.1 GBHO01041725 GBHO01038211 GBHO01034604 GBHO01034603 GBHO01034602 GBHO01034601 GBHO01034600 GBHO01034599 GBHO01034598 GBHO01034597 GBHO01034596 GBHO01001798 JAG01879.1 JAG05393.1 JAG09000.1 JAG09001.1 JAG09002.1 JAG09003.1 JAG09004.1 JAG09005.1 JAG09006.1 JAG09007.1 JAG09008.1 JAG41806.1 GALX01001116 JAB67350.1 GANO01004672 JAB55199.1 GGFK01006597 MBW39918.1 DS232085 EDS34366.1 GFDL01015905 JAV19140.1 KK852483 KDR22856.1 ATLV01023810 KE525347 KFB49652.1 KQ434896 KZC10728.1 APCN01003275 GDHF01030768 GDHF01029202 JAI21546.1 JAI23112.1 GAMC01002781 JAC03775.1 GL770938 EFZ09917.1 EZ000395 ACJ64269.1 LBMM01001560 KMQ96087.1 AAAB01008788 EAA03519.4 KZ288466 PBC25440.1 JR049862 AEY61102.1 KA645855 AFP60484.1 CH963849 EDW73973.1 ADMH02001585 ETN61967.1 KQ765605 OAD53870.1 GAKP01002491 JAC56461.1 GL443059 EFN62664.1 GGFM01003640 MBW24391.1 NEVH01012087 PNF30593.1 KQ435878 KOX70041.1 AXCM01009508 CP012524 ALC41482.1 UFQT01001564 UFQT01003550 SSX31025.1 SSX35107.1 GAMD01001195 JAB00396.1 GBXI01006034 JAD08258.1 CH940667 EDW57194.1 KQ414786 KOC60852.1 CM000158 EDW90012.1 CH933808 EDW08325.1 GDAI01001147 JAI16456.1 CH480816 EDW47376.1 CM000362 CM002911 EDX06598.1 KMY92955.1 AE013599 BT011056 KX531730 AAF58682.1 AAR31127.1 ANY27540.1 CH954179 EDV56502.1 KK107109 EZA59145.1 AXCN02001078 CH902619 EDV35553.1 GAPW01005986 JAC07612.1 JRES01000655 KNC29533.1 GL453466 EFN75977.1 QOIP01000008 RLU19194.1 GFDF01010946 JAV03138.1 AJWK01024813 GECZ01000405 JAS69364.1 GECU01013930 JAS93776.1 GEDC01010084 JAS27214.1 AK417533 BAN20748.1
KPI97929.1 KQ460883 KPJ11351.1 GAIX01013861 JAA78699.1 JTDY01002647 KOB71011.1 AGBW02012225 OWR45400.1 PYGN01002489 PSN30399.1 DQ440430 ABF18463.1 CH477828 EAT35860.1 APGK01057485 BT126689 KB741280 KB632033 AEE61651.1 ENN70944.1 ERL88184.1 LJIG01009094 KRT83767.1 CH479183 EDW36311.1 KQ971321 EFA00618.1 CM000071 EAL25834.1 JXUM01095302 GAPW01005525 GAPW01005524 KQ564187 JAC08073.1 KXJ72604.1 GFDG01001697 JAV17102.1 OUUW01000001 SPP74595.1 GBRD01013421 GBRD01013420 GBRD01013419 GDHC01020311 GDHC01014732 JAG52405.1 JAP98317.1 JAQ03897.1 CH916367 EDW02334.1 GBHO01041725 GBHO01038211 GBHO01034604 GBHO01034603 GBHO01034602 GBHO01034601 GBHO01034600 GBHO01034599 GBHO01034598 GBHO01034597 GBHO01034596 GBHO01001798 JAG01879.1 JAG05393.1 JAG09000.1 JAG09001.1 JAG09002.1 JAG09003.1 JAG09004.1 JAG09005.1 JAG09006.1 JAG09007.1 JAG09008.1 JAG41806.1 GALX01001116 JAB67350.1 GANO01004672 JAB55199.1 GGFK01006597 MBW39918.1 DS232085 EDS34366.1 GFDL01015905 JAV19140.1 KK852483 KDR22856.1 ATLV01023810 KE525347 KFB49652.1 KQ434896 KZC10728.1 APCN01003275 GDHF01030768 GDHF01029202 JAI21546.1 JAI23112.1 GAMC01002781 JAC03775.1 GL770938 EFZ09917.1 EZ000395 ACJ64269.1 LBMM01001560 KMQ96087.1 AAAB01008788 EAA03519.4 KZ288466 PBC25440.1 JR049862 AEY61102.1 KA645855 AFP60484.1 CH963849 EDW73973.1 ADMH02001585 ETN61967.1 KQ765605 OAD53870.1 GAKP01002491 JAC56461.1 GL443059 EFN62664.1 GGFM01003640 MBW24391.1 NEVH01012087 PNF30593.1 KQ435878 KOX70041.1 AXCM01009508 CP012524 ALC41482.1 UFQT01001564 UFQT01003550 SSX31025.1 SSX35107.1 GAMD01001195 JAB00396.1 GBXI01006034 JAD08258.1 CH940667 EDW57194.1 KQ414786 KOC60852.1 CM000158 EDW90012.1 CH933808 EDW08325.1 GDAI01001147 JAI16456.1 CH480816 EDW47376.1 CM000362 CM002911 EDX06598.1 KMY92955.1 AE013599 BT011056 KX531730 AAF58682.1 AAR31127.1 ANY27540.1 CH954179 EDV56502.1 KK107109 EZA59145.1 AXCN02001078 CH902619 EDV35553.1 GAPW01005986 JAC07612.1 JRES01000655 KNC29533.1 GL453466 EFN75977.1 QOIP01000008 RLU19194.1 GFDF01010946 JAV03138.1 AJWK01024813 GECZ01000405 JAS69364.1 GECU01013930 JAS93776.1 GEDC01010084 JAS27214.1 AK417533 BAN20748.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000245037
+ More
UP000008820 UP000019118 UP000030742 UP000008744 UP000007266 UP000001819 UP000069940 UP000249989 UP000095300 UP000268350 UP000001070 UP000069272 UP000192223 UP000002320 UP000027135 UP000030765 UP000076502 UP000075840 UP000075903 UP000075902 UP000076407 UP000075882 UP000036403 UP000075885 UP000007062 UP000095301 UP000242457 UP000005203 UP000007798 UP000000673 UP000075880 UP000000311 UP000235965 UP000075881 UP000053105 UP000075901 UP000075883 UP000092553 UP000008792 UP000053825 UP000002282 UP000192221 UP000009192 UP000076408 UP000001292 UP000000304 UP000000803 UP000008711 UP000053097 UP000075920 UP000075884 UP000075886 UP000007801 UP000037069 UP000008237 UP000279307 UP000092461
UP000008820 UP000019118 UP000030742 UP000008744 UP000007266 UP000001819 UP000069940 UP000249989 UP000095300 UP000268350 UP000001070 UP000069272 UP000192223 UP000002320 UP000027135 UP000030765 UP000076502 UP000075840 UP000075903 UP000075902 UP000076407 UP000075882 UP000036403 UP000075885 UP000007062 UP000095301 UP000242457 UP000005203 UP000007798 UP000000673 UP000075880 UP000000311 UP000235965 UP000075881 UP000053105 UP000075901 UP000075883 UP000092553 UP000008792 UP000053825 UP000002282 UP000192221 UP000009192 UP000076408 UP000001292 UP000000304 UP000000803 UP000008711 UP000053097 UP000075920 UP000075884 UP000075886 UP000007801 UP000037069 UP000008237 UP000279307 UP000092461
Pfam
PF00160 Pro_isomerase
Interpro
SUPFAM
SSF50891
SSF50891
Gene 3D
ProteinModelPortal
H9ISX2
A0A1E1VXF9
A0A2H1X3Q6
A0A194PYV6
A0A194R660
S4NNN1
+ More
A0A0L7L6R7 A0A212EV77 A0A2P8XEH7 Q1HQL4 Q16ND9 J3JTX9 A0A0T6BA50 B4GIJ1 D6WH15 Q28YY1 A0A023EF95 A0A1I8Q6D8 A0A1L8EEG4 A0A3B0J177 A0A0K8SHZ8 B4J8H5 A0A0A9WNK3 A0A182FZR7 V5GB57 A0A1W4WSC5 U5EM09 A0A2M4AH27 B0WTC8 A0A1Q3EV05 A0A067RG54 A0A084WHF8 A0A154PHD2 A0A2C9GR40 A0A182ULT8 A0A182TGZ0 A0A182WZX8 A0A182KSY7 A0A0K8U8X5 W8CD06 E9JB68 B8RIX1 A0A0J7KZY8 A0A182P1S3 Q7QKM6 A0A1I8MFE8 A0A2A3E166 V9IKC9 A0A088AA90 T1PCI8 B4MPD4 W5JCB2 A0A182JLB4 A0A310SJU3 A0A034WLK7 E2AVB0 A0A2M3Z7A8 A0A2J7QPV5 A0A182K8H2 A0A0N0BDF9 A0A182T6G9 A0A182MW09 A0A0M4EV79 A0A336MMT1 T1DQE9 A0A0A1XCS5 B4MFM3 A0A0L7QQF9 B4P7H7 A0A1W4UGG9 B4KRQ5 A0A182Y217 A0A0K8TQL5 B4HNE0 B4QB65 Q6NP72 B3NSK0 A0A026WSX4 A0A182W9D7 A0A182N2L5 A0A182QVD3 B3MET9 A0A023EE51 A0A0L0CAU8 E2C7U2 A0A3L8DFM3 A0A1L8D9J2 A0A1B0CRH9 A0A1B6H3Y2 A0A1B6J3K9 A0A1B6DNG9 R4WDB8
A0A0L7L6R7 A0A212EV77 A0A2P8XEH7 Q1HQL4 Q16ND9 J3JTX9 A0A0T6BA50 B4GIJ1 D6WH15 Q28YY1 A0A023EF95 A0A1I8Q6D8 A0A1L8EEG4 A0A3B0J177 A0A0K8SHZ8 B4J8H5 A0A0A9WNK3 A0A182FZR7 V5GB57 A0A1W4WSC5 U5EM09 A0A2M4AH27 B0WTC8 A0A1Q3EV05 A0A067RG54 A0A084WHF8 A0A154PHD2 A0A2C9GR40 A0A182ULT8 A0A182TGZ0 A0A182WZX8 A0A182KSY7 A0A0K8U8X5 W8CD06 E9JB68 B8RIX1 A0A0J7KZY8 A0A182P1S3 Q7QKM6 A0A1I8MFE8 A0A2A3E166 V9IKC9 A0A088AA90 T1PCI8 B4MPD4 W5JCB2 A0A182JLB4 A0A310SJU3 A0A034WLK7 E2AVB0 A0A2M3Z7A8 A0A2J7QPV5 A0A182K8H2 A0A0N0BDF9 A0A182T6G9 A0A182MW09 A0A0M4EV79 A0A336MMT1 T1DQE9 A0A0A1XCS5 B4MFM3 A0A0L7QQF9 B4P7H7 A0A1W4UGG9 B4KRQ5 A0A182Y217 A0A0K8TQL5 B4HNE0 B4QB65 Q6NP72 B3NSK0 A0A026WSX4 A0A182W9D7 A0A182N2L5 A0A182QVD3 B3MET9 A0A023EE51 A0A0L0CAU8 E2C7U2 A0A3L8DFM3 A0A1L8D9J2 A0A1B0CRH9 A0A1B6H3Y2 A0A1B6J3K9 A0A1B6DNG9 R4WDB8
Ontologies
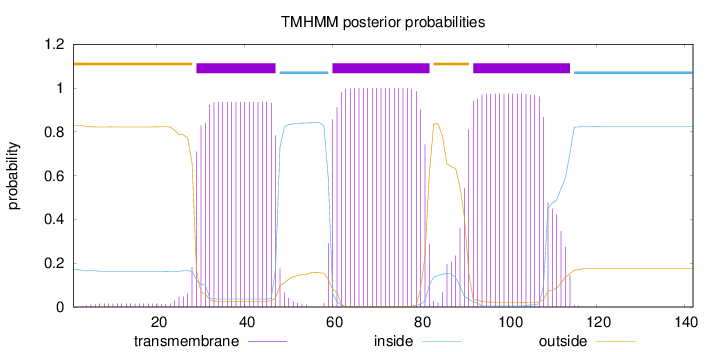
Topology
Length:
142
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
61.16985
Exp number, first 60 AAs:
19.39643
Total prob of N-in:
0.17132
POSSIBLE N-term signal
sequence
outside
1 - 28
TMhelix
29 - 47
inside
48 - 59
TMhelix
60 - 82
outside
83 - 91
TMhelix
92 - 114
inside
115 - 142
Population Genetic Test Statistics
Pi
295.848229
Theta
235.79539
Tajima's D
0.81822
CLR
0.002795
CSRT
0.607469626518674
Interpretation
Uncertain
