Gene
KWMTBOMO13166 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000347
Annotation
PREDICTED:_protein_lethal(3)malignant_blood_neoplasm_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.993
Sequence
CDS
ATGGATCTACTCAGAAAGATTAATATTATTGTGGTGATTACTACATGCATATTACTCCAAGCAGAAGGAGCCGACAAATATACTGATGAGAAAAGACCGTACGAATTCGGTTTCACGATTGACGGGCAGCAGCATAGACACGAAAAGAAAGATAAGAACGGCATTATCATGGGAGAGTTCGGATTCATCACTGCCGATGATGTATATCATGTGACAGTGTACGCTACTGACGAAAATGGAAGATTCAAAATAATTTCTATGAAGAACATACATTTGAAATCACAATCGACATCGCAAAGACAGGGTCACTCGTTGCAAATACCGAACACACAAAACGCTATAGCGTCAACTCAATCGCCCACCCAAAAACCAGAAAACACCCTCAAATCTATATCTATAGAACCACCAGCTAAATCCTGTTCGCATTGCAGTCTGCCGACAACTTCTACAGTTGCTCCGGCATCTGCTAAACCAGCATTTATCAATGGTGGAATTGGCGGCACAAAATCGGACGTACAAAATAATCAAGTGCCAACTTACAATGTAAAACAAAACAATGCTGATTCACAATACCCTCAACAAGGAGATTTCAAAAACTTCCCAGGTCAACAACTCAATCCTCAAAATGTAGGAAACCAACAAACTCCTGACGTACAAAATAGCCGTACAAATCAAAATAATAAACAAACACAAGGGGCTTCTGGAAGCTTCCTACCACTGAGTGGTAGTGGAGGCTCTCAAAATGGTTTTAAGCAAACCCCTGTCCAAGCCCTGTCTACAATAGCGAACCAAGGTGGAGACAATCCAGAATACCCAACTGGACTAAGGAATAGCTTTGATTCTCCACAAGGACAACAAAATGAATTCGGTGCTCCACAGAGACAACAAAACAGCTTTGGTGCACCACAAGGAAATCAAAATGGATTCGGTGCTCCACATGCTGCCGCTGAAGGCTTAGGAACTTCCAAGGGATCATCTGATAGTAATATTTTCAACAACCAAAATTCAAAAACACCTAATTCATTTAATCAGGGCTCGGATGAACAAAGAGGAACACGTGATGAACAAACGGGTAGCAACGCTGGCGGTACAGCTAATGAAAGAATTAATAGTATTAACGCAGGTCAAACTACTACTGGGGCTGTTGCAGGATCTCGTTCACCTAAGCTGTACAATGTAAAACCAGATCAATCTTCCAATGGCCTGAGCCCAAGAGAACAGTCTCGATTCGGACAAGCCAGTGGCAATAAAATTAGACCAGAAAATGGTCAATCTAACAACGAGAAACCGACATTGACATACGCTCAAATCCAAATTGTTGATAAGAATACTGACATTCACTCCAAACGCCCTGGAGAAAAAGACGGATTACCTAACGGTATTACCGAAAACGACATGGTTCATCTTCTTTATACGTTCAATTACACTTTAGGTTTTCAAGCACATTTTGAAGAAGGATACTCGAACGGCGTCAAACAAGGATACTATTACGTTACAGGACGAAATGGAATCAGAACAAGGATCGATTACATAGCAGACGATAAAGGCTTTAGACCTAAAATATCCCAAGAGGTGCTTGATTTACTGTCTGAAGATGTGCCGAAACCTGAAACAGAAAAAGACGAAAAATACGGTCTGAAGGGCTATGAATTTAAATGGTTATATTATCCTCTCGAGTCTAAAGCGCGGTAG
Protein
MDLLRKINIIVVITTCILLQAEGADKYTDEKRPYEFGFTIDGQQHRHEKKDKNGIIMGEFGFITADDVYHVTVYATDENGRFKIISMKNIHLKSQSTSQRQGHSLQIPNTQNAIASTQSPTQKPENTLKSISIEPPAKSCSHCSLPTTSTVAPASAKPAFINGGIGGTKSDVQNNQVPTYNVKQNNADSQYPQQGDFKNFPGQQLNPQNVGNQQTPDVQNSRTNQNNKQTQGASGSFLPLSGSGGSQNGFKQTPVQALSTIANQGGDNPEYPTGLRNSFDSPQGQQNEFGAPQRQQNSFGAPQGNQNGFGAPHAAAEGLGTSKGSSDSNIFNNQNSKTPNSFNQGSDEQRGTRDEQTGSNAGGTANERINSINAGQTTTGAVAGSRSPKLYNVKPDQSSNGLSPREQSRFGQASGNKIRPENGQSNNEKPTLTYAQIQIVDKNTDIHSKRPGEKDGLPNGITENDMVHLLYTFNYTLGFQAHFEEGYSNGVKQGYYYVTGRNGIRTRIDYIADDKGFRPKISQEVLDLLSEDVPKPETEKDEKYGLKGYEFKWLYYPLESKAR
Summary
Uniprot
Proteomes
PRIDE
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
Ontologies
GO
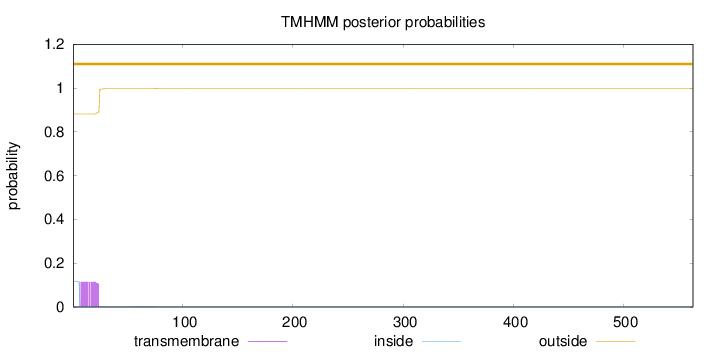
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.953077
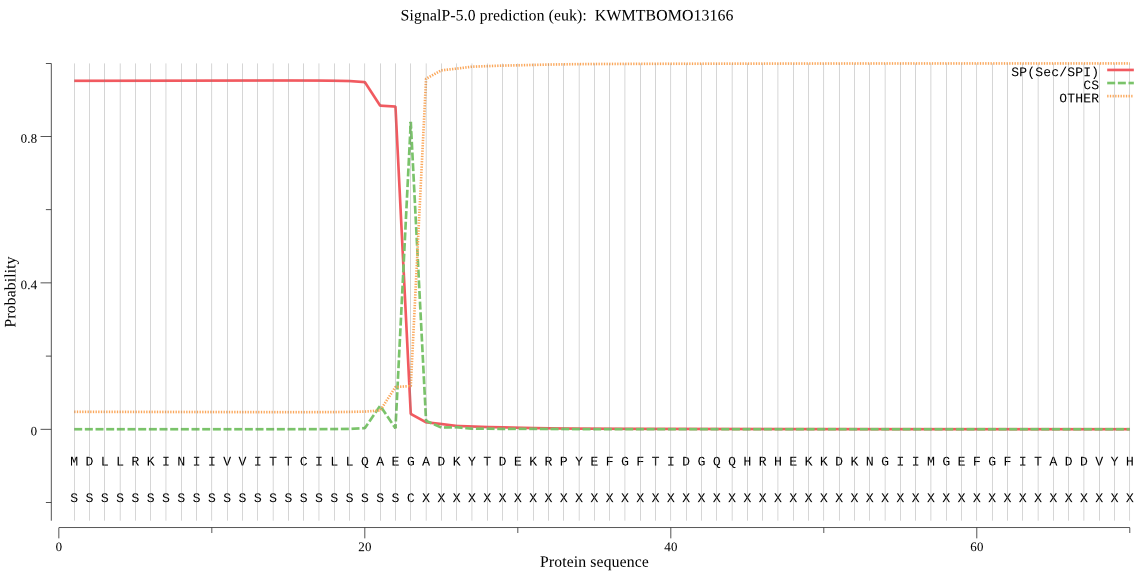
Length:
563
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.10895
Exp number, first 60 AAs:
2.0738
Total prob of N-in:
0.11802
outside
1 - 563
Population Genetic Test Statistics
Pi
230.172571
Theta
189.079828
Tajima's D
0.957758
CLR
0
CSRT
0.645767711614419
Interpretation
Uncertain
