Gene
KWMTBOMO13165 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000346
Annotation
cuticular_protein_RR-1_motif_23_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.64
Sequence
CDS
ATGCTTAAATACGTGACAGTCGCTTGCGTGCTGGTAGCCCTTTGCTCAGGTGCTCCACAACAAAACCCCCAAGATGTTCAGATCCTACGTTTTGACAGCAACGTGGAACCCGATGGCTACAGCTTTGCGTACGAAACTAGTGATGGTACATCCAGACAAGAAGAAGGCAAACTCGACAACCCACAGTCCGAGAATGCAGCTCTGACAGTTACTGGACAGTATGCTTACGTAGCTCCTGACGGCAAGCACTACACTGTCACTTTCACAGCGGGACCCAACGGCTTCCAACCCAAAACCTCATTGGGCCAGAAGTAA
Protein
MLKYVTVACVLVALCSGAPQQNPQDVQILRFDSNVEPDGYSFAYETSDGTSRQEEGKLDNPQSENAALTVTGQYAYVAPDGKHYTVTFTAGPNGFQPKTSLGQK
Summary
Uniprot
C0H6L4
G8FVQ5
A0A212ESP2
A0A1E1W6B4
A0A194PY21
A0A194R0X8
+ More
A0A2H1VJA8 A0A2A4IU70 A0A0L7H1F6 A0A1L8E3C3 A0A336LX70 A0A1B0GNX6 W8BQN7 I4DJF4 T1DQI3 A0A182FCP1 A0A1W4VAI2 A0A0A1XHQ1 C0H6M2 A0A3B0JXI1 W8BXU7 W5J9B1 A0A034V1C0 A0A034V1Z8 A0A0K8W0S8 A0A1J1IRW4 H9ISW2 D6WMB2 A0A2H4QYZ4 B3NGE3 A0A2H4QZ11 A0A2H4QYX4 A0A2H4QYZ2 A0A2H4QYY0 A0A2H4QZ41 B4LHF0 A0A2H4QZ04 A0A194R1G9 A0A034WSW4 A0A2H4QYX0 A0A2H1VIV9 A0A0K8U9Z8 T1PAJ5 A0A194PY16 A0A2A4ITS3 A0A2H4QYX6 A0A3B0KMU2 P91939 B4IZT2 A0A067RF26 A0A182Q833 W8BY71 A0A0M4EHM0 Q8IGZ8 A0A0M4EYK3 A0A3S2NUQ1 A0A0A1XNL9 A0A023EEV7 N6TE67 A0A1Q3FIR8 B4IZT5 A0A1J1IRY0 B4IZU0 D6WMB0 D6WMB3 A0A1I8NP48 B4ML04 B4IZT6 B4KWN7 B0XA27 A0A1I8MMH9 B4LHE8 A0A1I8M2I6 B4KWM2 B4ML00 Q2TAR4 B3NGE1 Q2LZE0 B4H6H1 A0A182TZI8 B4HUQ4 B4QJA5 A0A3B0KFM6 A0A1B0G041 A0A1A9XR09 A0A1B0C206 B4PJD4 A0A0K8V4L5 Q16IX7 B4KWP2 B4PJE2 A0A1I8JTT8 A0A182KPF5 Q7PNS3 A0A2H4QYV9 P92181 A0A1W4VAJ2 B4MKZ8
A0A2H1VJA8 A0A2A4IU70 A0A0L7H1F6 A0A1L8E3C3 A0A336LX70 A0A1B0GNX6 W8BQN7 I4DJF4 T1DQI3 A0A182FCP1 A0A1W4VAI2 A0A0A1XHQ1 C0H6M2 A0A3B0JXI1 W8BXU7 W5J9B1 A0A034V1C0 A0A034V1Z8 A0A0K8W0S8 A0A1J1IRW4 H9ISW2 D6WMB2 A0A2H4QYZ4 B3NGE3 A0A2H4QZ11 A0A2H4QYX4 A0A2H4QYZ2 A0A2H4QYY0 A0A2H4QZ41 B4LHF0 A0A2H4QZ04 A0A194R1G9 A0A034WSW4 A0A2H4QYX0 A0A2H1VIV9 A0A0K8U9Z8 T1PAJ5 A0A194PY16 A0A2A4ITS3 A0A2H4QYX6 A0A3B0KMU2 P91939 B4IZT2 A0A067RF26 A0A182Q833 W8BY71 A0A0M4EHM0 Q8IGZ8 A0A0M4EYK3 A0A3S2NUQ1 A0A0A1XNL9 A0A023EEV7 N6TE67 A0A1Q3FIR8 B4IZT5 A0A1J1IRY0 B4IZU0 D6WMB0 D6WMB3 A0A1I8NP48 B4ML04 B4IZT6 B4KWN7 B0XA27 A0A1I8MMH9 B4LHE8 A0A1I8M2I6 B4KWM2 B4ML00 Q2TAR4 B3NGE1 Q2LZE0 B4H6H1 A0A182TZI8 B4HUQ4 B4QJA5 A0A3B0KFM6 A0A1B0G041 A0A1A9XR09 A0A1B0C206 B4PJD4 A0A0K8V4L5 Q16IX7 B4KWP2 B4PJE2 A0A1I8JTT8 A0A182KPF5 Q7PNS3 A0A2H4QYV9 P92181 A0A1W4VAJ2 B4MKZ8
Pubmed
19121390
19280704
22118469
26354079
26227816
24495485
+ More
22651552 25830018 20920257 23761445 25348373 18362917 19820115 17994087 25315136 9383064 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 24945155 23537049 15632085 22936249 17550304 17510324 20966253 12364791 14747013 17210077
22651552 25830018 20920257 23761445 25348373 18362917 19820115 17994087 25315136 9383064 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 24945155 23537049 15632085 22936249 17550304 17510324 20966253 12364791 14747013 17210077
EMBL
BABH01013041
BR000524
FAA00525.1
JN082725
AER27822.1
AGBW02012745
+ More
OWR44512.1 GDQN01008534 JAT82520.1 KQ459586 KPI97923.1 KQ460883 KPJ11357.1 ODYU01002880 SOQ40913.1 NWSH01008139 PCG62680.1 JTDY01023902 KOB14831.1 GFDF01000863 JAV13221.1 UFQT01000259 SSX22515.1 AJVK01014117 GAMC01002990 JAC03566.1 AK401422 BAM18044.1 GAMD01001140 JAB00451.1 GBXI01003413 JAD10879.1 BR000531 FAA00533.1 OUUW01000009 SPP85122.1 GAMC01000330 JAC06226.1 ADMH02002086 ETN59435.1 GAKP01023378 JAC35580.1 GAKP01023380 JAC35578.1 GDHF01007581 JAI44733.1 CVRI01000059 CRL02971.1 BABH01013046 BABH01013047 KQ971343 EFA04245.1 KY066650 ATY36878.1 CH954178 EDV51109.1 KY066651 ATY36879.1 KY066629 KY066631 KY066634 ATY36857.1 ATY36859.1 ATY36862.1 KY066648 KY066649 KY066652 KY066653 KY066654 KY066655 KY066656 ATY36876.1 ATY36877.1 ATY36880.1 ATY36881.1 ATY36882.1 ATY36883.1 ATY36884.1 KY066638 KY066639 KY066640 KY066642 KY066643 KY066644 KY066645 KY066646 KY066647 ATY36866.1 ATY36867.1 ATY36868.1 ATY36870.1 ATY36871.1 ATY36872.1 ATY36873.1 ATY36874.1 ATY36875.1 KY066633 ATY36861.1 CH940647 EDW68480.1 KY066641 ATY36869.1 KPJ11364.1 GAKP01000301 JAC58651.1 KY066637 ATY36865.1 ODYU01002809 SOQ40763.1 GDHF01028822 JAI23492.1 KA645719 AFP60348.1 KPI97918.1 NWSH01007118 PCG63079.1 KY066630 ATY36858.1 SPP85128.1 U83247 AE014296 KX531463 AAB88063.1 AAF50688.1 AGB94161.1 ANY27273.1 CH916366 EDV95667.1 KK852675 KDR18706.1 AXCN02000482 GAMC01005012 JAC01544.1 CP012525 ALC43214.1 BT001515 AAN71270.1 ALC43600.1 RSAL01000175 RVE45046.1 GBXI01001383 JAD12909.1 GAPW01006364 JAC07234.1 APGK01032633 KB740848 ENN78644.1 GFDL01007568 JAV27477.1 EDV95670.1 CRL02975.1 EDV95675.1 EFA04243.1 EFA04246.1 CH963847 EDW72929.1 EDV95671.1 CH933809 EDW17484.1 DS232561 EDS43453.1 EDW68478.1 EDW17469.1 EDW72925.1 BC110760 AAI10761.1 EDV51107.1 CH379069 EAL29569.1 CH479214 EDW33399.1 CH480817 EDW50675.1 CM000363 CM002912 EDX09414.1 KMY97874.1 SPP85129.1 CCAG010009302 JXJN01024293 CM000159 EDW93602.1 GDHF01018475 JAI33839.1 CH478046 EAT34219.1 EDW17489.1 EDW93610.2 APCN01000091 AAAB01008960 EAA11701.4 KY066636 ATY36864.1 U84755 U84745 AY071303 KX532141 AAB48462.1 AAB88064.1 AAF50687.1 AAL48925.1 ANY27951.1 EDW72923.1
OWR44512.1 GDQN01008534 JAT82520.1 KQ459586 KPI97923.1 KQ460883 KPJ11357.1 ODYU01002880 SOQ40913.1 NWSH01008139 PCG62680.1 JTDY01023902 KOB14831.1 GFDF01000863 JAV13221.1 UFQT01000259 SSX22515.1 AJVK01014117 GAMC01002990 JAC03566.1 AK401422 BAM18044.1 GAMD01001140 JAB00451.1 GBXI01003413 JAD10879.1 BR000531 FAA00533.1 OUUW01000009 SPP85122.1 GAMC01000330 JAC06226.1 ADMH02002086 ETN59435.1 GAKP01023378 JAC35580.1 GAKP01023380 JAC35578.1 GDHF01007581 JAI44733.1 CVRI01000059 CRL02971.1 BABH01013046 BABH01013047 KQ971343 EFA04245.1 KY066650 ATY36878.1 CH954178 EDV51109.1 KY066651 ATY36879.1 KY066629 KY066631 KY066634 ATY36857.1 ATY36859.1 ATY36862.1 KY066648 KY066649 KY066652 KY066653 KY066654 KY066655 KY066656 ATY36876.1 ATY36877.1 ATY36880.1 ATY36881.1 ATY36882.1 ATY36883.1 ATY36884.1 KY066638 KY066639 KY066640 KY066642 KY066643 KY066644 KY066645 KY066646 KY066647 ATY36866.1 ATY36867.1 ATY36868.1 ATY36870.1 ATY36871.1 ATY36872.1 ATY36873.1 ATY36874.1 ATY36875.1 KY066633 ATY36861.1 CH940647 EDW68480.1 KY066641 ATY36869.1 KPJ11364.1 GAKP01000301 JAC58651.1 KY066637 ATY36865.1 ODYU01002809 SOQ40763.1 GDHF01028822 JAI23492.1 KA645719 AFP60348.1 KPI97918.1 NWSH01007118 PCG63079.1 KY066630 ATY36858.1 SPP85128.1 U83247 AE014296 KX531463 AAB88063.1 AAF50688.1 AGB94161.1 ANY27273.1 CH916366 EDV95667.1 KK852675 KDR18706.1 AXCN02000482 GAMC01005012 JAC01544.1 CP012525 ALC43214.1 BT001515 AAN71270.1 ALC43600.1 RSAL01000175 RVE45046.1 GBXI01001383 JAD12909.1 GAPW01006364 JAC07234.1 APGK01032633 KB740848 ENN78644.1 GFDL01007568 JAV27477.1 EDV95670.1 CRL02975.1 EDV95675.1 EFA04243.1 EFA04246.1 CH963847 EDW72929.1 EDV95671.1 CH933809 EDW17484.1 DS232561 EDS43453.1 EDW68478.1 EDW17469.1 EDW72925.1 BC110760 AAI10761.1 EDV51107.1 CH379069 EAL29569.1 CH479214 EDW33399.1 CH480817 EDW50675.1 CM000363 CM002912 EDX09414.1 KMY97874.1 SPP85129.1 CCAG010009302 JXJN01024293 CM000159 EDW93602.1 GDHF01018475 JAI33839.1 CH478046 EAT34219.1 EDW17489.1 EDW93610.2 APCN01000091 AAAB01008960 EAA11701.4 KY066636 ATY36864.1 U84755 U84745 AY071303 KX532141 AAB48462.1 AAB88064.1 AAF50687.1 AAL48925.1 ANY27951.1 EDW72923.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000037510
+ More
UP000092462 UP000069272 UP000192221 UP000268350 UP000000673 UP000183832 UP000007266 UP000008711 UP000008792 UP000095301 UP000000803 UP000001070 UP000027135 UP000075886 UP000092553 UP000283053 UP000019118 UP000095300 UP000007798 UP000009192 UP000002320 UP000001819 UP000008744 UP000075902 UP000001292 UP000000304 UP000092444 UP000092443 UP000092460 UP000002282 UP000008820 UP000075840 UP000075882 UP000007062
UP000092462 UP000069272 UP000192221 UP000268350 UP000000673 UP000183832 UP000007266 UP000008711 UP000008792 UP000095301 UP000000803 UP000001070 UP000027135 UP000075886 UP000092553 UP000283053 UP000019118 UP000095300 UP000007798 UP000009192 UP000002320 UP000001819 UP000008744 UP000075902 UP000001292 UP000000304 UP000092444 UP000092443 UP000092460 UP000002282 UP000008820 UP000075840 UP000075882 UP000007062
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6L4
G8FVQ5
A0A212ESP2
A0A1E1W6B4
A0A194PY21
A0A194R0X8
+ More
A0A2H1VJA8 A0A2A4IU70 A0A0L7H1F6 A0A1L8E3C3 A0A336LX70 A0A1B0GNX6 W8BQN7 I4DJF4 T1DQI3 A0A182FCP1 A0A1W4VAI2 A0A0A1XHQ1 C0H6M2 A0A3B0JXI1 W8BXU7 W5J9B1 A0A034V1C0 A0A034V1Z8 A0A0K8W0S8 A0A1J1IRW4 H9ISW2 D6WMB2 A0A2H4QYZ4 B3NGE3 A0A2H4QZ11 A0A2H4QYX4 A0A2H4QYZ2 A0A2H4QYY0 A0A2H4QZ41 B4LHF0 A0A2H4QZ04 A0A194R1G9 A0A034WSW4 A0A2H4QYX0 A0A2H1VIV9 A0A0K8U9Z8 T1PAJ5 A0A194PY16 A0A2A4ITS3 A0A2H4QYX6 A0A3B0KMU2 P91939 B4IZT2 A0A067RF26 A0A182Q833 W8BY71 A0A0M4EHM0 Q8IGZ8 A0A0M4EYK3 A0A3S2NUQ1 A0A0A1XNL9 A0A023EEV7 N6TE67 A0A1Q3FIR8 B4IZT5 A0A1J1IRY0 B4IZU0 D6WMB0 D6WMB3 A0A1I8NP48 B4ML04 B4IZT6 B4KWN7 B0XA27 A0A1I8MMH9 B4LHE8 A0A1I8M2I6 B4KWM2 B4ML00 Q2TAR4 B3NGE1 Q2LZE0 B4H6H1 A0A182TZI8 B4HUQ4 B4QJA5 A0A3B0KFM6 A0A1B0G041 A0A1A9XR09 A0A1B0C206 B4PJD4 A0A0K8V4L5 Q16IX7 B4KWP2 B4PJE2 A0A1I8JTT8 A0A182KPF5 Q7PNS3 A0A2H4QYV9 P92181 A0A1W4VAJ2 B4MKZ8
A0A2H1VJA8 A0A2A4IU70 A0A0L7H1F6 A0A1L8E3C3 A0A336LX70 A0A1B0GNX6 W8BQN7 I4DJF4 T1DQI3 A0A182FCP1 A0A1W4VAI2 A0A0A1XHQ1 C0H6M2 A0A3B0JXI1 W8BXU7 W5J9B1 A0A034V1C0 A0A034V1Z8 A0A0K8W0S8 A0A1J1IRW4 H9ISW2 D6WMB2 A0A2H4QYZ4 B3NGE3 A0A2H4QZ11 A0A2H4QYX4 A0A2H4QYZ2 A0A2H4QYY0 A0A2H4QZ41 B4LHF0 A0A2H4QZ04 A0A194R1G9 A0A034WSW4 A0A2H4QYX0 A0A2H1VIV9 A0A0K8U9Z8 T1PAJ5 A0A194PY16 A0A2A4ITS3 A0A2H4QYX6 A0A3B0KMU2 P91939 B4IZT2 A0A067RF26 A0A182Q833 W8BY71 A0A0M4EHM0 Q8IGZ8 A0A0M4EYK3 A0A3S2NUQ1 A0A0A1XNL9 A0A023EEV7 N6TE67 A0A1Q3FIR8 B4IZT5 A0A1J1IRY0 B4IZU0 D6WMB0 D6WMB3 A0A1I8NP48 B4ML04 B4IZT6 B4KWN7 B0XA27 A0A1I8MMH9 B4LHE8 A0A1I8M2I6 B4KWM2 B4ML00 Q2TAR4 B3NGE1 Q2LZE0 B4H6H1 A0A182TZI8 B4HUQ4 B4QJA5 A0A3B0KFM6 A0A1B0G041 A0A1A9XR09 A0A1B0C206 B4PJD4 A0A0K8V4L5 Q16IX7 B4KWP2 B4PJE2 A0A1I8JTT8 A0A182KPF5 Q7PNS3 A0A2H4QYV9 P92181 A0A1W4VAJ2 B4MKZ8
Ontologies
GO
Topology
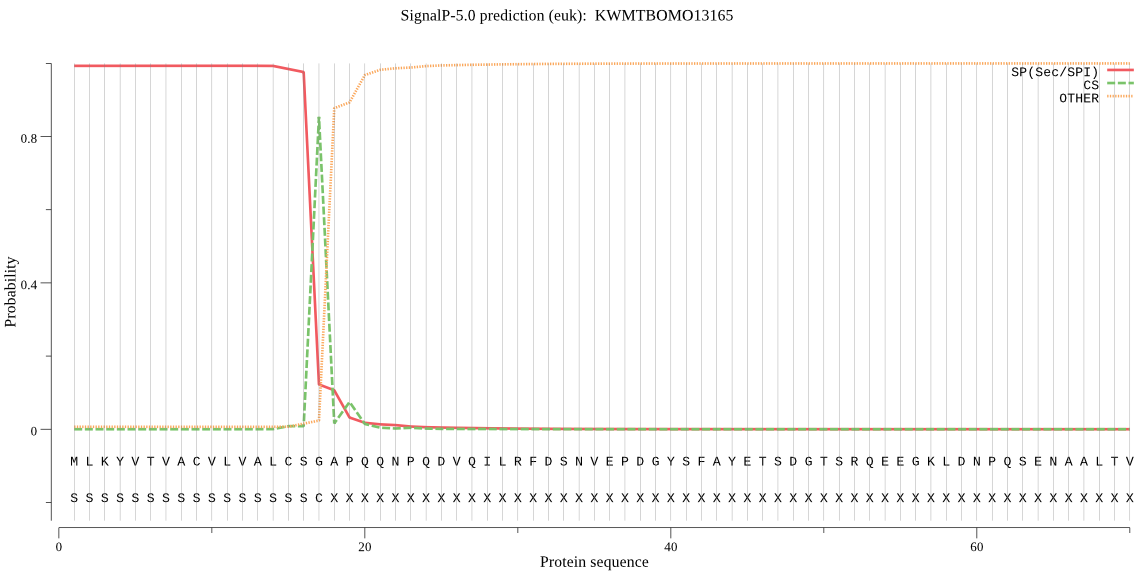
SignalP
Position: 1 - 17,
Likelihood: 0.993014
Length:
104
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0988100000000001
Exp number, first 60 AAs:
0.09797
Total prob of N-in:
0.38689
outside
1 - 104

Population Genetic Test Statistics
Pi
388.210545
Theta
228.829446
Tajima's D
2.184813
CLR
0.321127
CSRT
0.908404579771011
Interpretation
Uncertain
