Gene
KWMTBOMO13152 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000337
Annotation
PREDICTED:_cuticular_protein_RR-1_motif_33_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.259 Extracellular Reliability : 1.181
Sequence
CDS
ATGATCTCAATACTGTTGTGCACATTGGCAGTGTTTTACGCAGTCCATGCGCAACAACATTCAATTAATGATCCCATTCCCATCATACGATATGAATCTGATGGACCTAATCCCGATGGATCCTACAAATGGCTATATGAAACCGGAAATGAAATAAACGCTGAAGAGACTGGCTACGTGAAAAACTTCGGTAAAGGAGAAGGTGAAGAAGTGCAGGTAGCTGAAGGAAAATTCAGCTACAAAGCACCTGATGGTTCGCTAATTGCGCTTTCATATATCGCCGATGAAAATGGTTTTCAACCTCAGGTATTATAA
Protein
MISILLCTLAVFYAVHAQQHSINDPIPIIRYESDGPNPDGSYKWLYETGNEINAEETGYVKNFGKGEGEEVQVAEGKFSYKAPDGSLIALSYIADENGFQPQVL
Summary
Uniprot
C0H6M5
A0A2H1V805
A0A0L7L4J5
A0A212FBE3
A0A3S2NQ74
I4DKM8
+ More
A0A194PY11 A0A194R1H4 A0A224XRL8 R4G543 T1I0T8 A0A2R7WSK8 A0A2S1ZS73 R4G3C1 T1I4C4 A0A0T6BG89 A0A171ALV7 A0A146MF21 A0A0A9XU59 Q17G30 A0A2P8Y0M8 A0A2J7R701 A0A194Q3B6 A0A2H1VDC4 A0A182KCU6 A0A2A4J2S0 A0A2A4IU20 A0A0A9ZEM1 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 T1I0T7 A0A182IU01 R4FJB9 A0A146L0M8 A0A139WHQ8 A0A182QGA7 A0A182MIF1 A0A212F0I8 A0A194R101 A0A1B0DG98 A0A182TXD1 D6WNV0 A0A1L8E3U5 A0A182HRK7 A0A182NUJ4 A0A182PJH7 A0A182RHG2 A0A088A4T5 Q9BPR1 A0A2H8TQ63 T1GT21 A0A1S4N1V2 A0A232FDH9 K7J4G1 A0A182WGU5 A0A1W4XP64 A0A0Q5VPK8 A0A2R7WPG2 F4X6E7 A0A3Q8HGL4 E0W3K9 A0A1B0GJ98 A0A310SI56 A0A182YDM7 A0A1W4XNY4 B4LHG0 A0A1W4U635 A0A195D5A8 A0A3B0JXJ0 A0A1B6LWW6 A0A1W4XN34 A0A182F693 A0A1I8NPH7 T1P8T8 A0A154P7J2 A0A2A3EGY6 A0A0N1IU57 A0A2S2PNN1 Q2LZE7 A0A0M8ZQ66 W5JPT7 B3MA08 B4PJE5 A0A0J9RAX5 B4P5C3 B4KWN0 E9IFU8 A8DRW0 A0A1W4XP74 A0A151X6J9 A0A1J1ILD8 B4HPA2 A0A0L7QVZ5 B4QCU1 C4WUY0 E2BGZ7 A0A0L7QW49
A0A194PY11 A0A194R1H4 A0A224XRL8 R4G543 T1I0T8 A0A2R7WSK8 A0A2S1ZS73 R4G3C1 T1I4C4 A0A0T6BG89 A0A171ALV7 A0A146MF21 A0A0A9XU59 Q17G30 A0A2P8Y0M8 A0A2J7R701 A0A194Q3B6 A0A2H1VDC4 A0A182KCU6 A0A2A4J2S0 A0A2A4IU20 A0A0A9ZEM1 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 T1I0T7 A0A182IU01 R4FJB9 A0A146L0M8 A0A139WHQ8 A0A182QGA7 A0A182MIF1 A0A212F0I8 A0A194R101 A0A1B0DG98 A0A182TXD1 D6WNV0 A0A1L8E3U5 A0A182HRK7 A0A182NUJ4 A0A182PJH7 A0A182RHG2 A0A088A4T5 Q9BPR1 A0A2H8TQ63 T1GT21 A0A1S4N1V2 A0A232FDH9 K7J4G1 A0A182WGU5 A0A1W4XP64 A0A0Q5VPK8 A0A2R7WPG2 F4X6E7 A0A3Q8HGL4 E0W3K9 A0A1B0GJ98 A0A310SI56 A0A182YDM7 A0A1W4XNY4 B4LHG0 A0A1W4U635 A0A195D5A8 A0A3B0JXJ0 A0A1B6LWW6 A0A1W4XN34 A0A182F693 A0A1I8NPH7 T1P8T8 A0A154P7J2 A0A2A3EGY6 A0A0N1IU57 A0A2S2PNN1 Q2LZE7 A0A0M8ZQ66 W5JPT7 B3MA08 B4PJE5 A0A0J9RAX5 B4P5C3 B4KWN0 E9IFU8 A8DRW0 A0A1W4XP74 A0A151X6J9 A0A1J1ILD8 B4HPA2 A0A0L7QVZ5 B4QCU1 C4WUY0 E2BGZ7 A0A0L7QW49
Pubmed
19121390
19280704
26227816
22118469
22651552
26354079
+ More
29712872 26823975 25401762 17510324 29403074 20966253 12364791 14747013 17210077 18362917 19820115 11483438 20566863 28648823 20075255 17994087 21719571 25244985 25315136 15632085 20920257 23761445 17550304 22936249 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20798317
29712872 26823975 25401762 17510324 29403074 20966253 12364791 14747013 17210077 18362917 19820115 11483438 20566863 28648823 20075255 17994087 21719571 25244985 25315136 15632085 20920257 23761445 17550304 22936249 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20798317
EMBL
BABH01013052
BR000534
FAA00536.1
ODYU01001157
SOQ36975.1
JTDY01002969
+ More
KOB70397.1 AGBW02009345 OWR51056.1 RSAL01000156 RVE45634.1 AK401846 BAM18468.1 KQ459586 KPI97913.1 KQ460883 KPJ11369.1 GFTR01002672 JAW13754.1 GAHY01000221 JAA77289.1 ACPB03017958 ACPB03017959 KK855351 PTY22120.1 MF942789 AWK28312.1 GAHY01002330 JAA75180.1 ACPB03009497 LJIG01000626 KRT86314.1 GEMB01000898 JAS02243.1 GDHC01001027 JAQ17602.1 GBHO01020110 JAG23494.1 CH477267 EAT45440.1 PYGN01001077 PSN37817.1 NEVH01006738 PNF36609.1 KPI97900.1 ODYU01001934 SOQ38827.1 NWSH01003794 PCG65814.1 NWSH01007872 PCG62774.1 GBHO01003344 JAG40260.1 AAAB01008980 EAA14379.5 ACPB03017960 GAHY01002338 JAA75172.1 GDHC01017817 JAQ00812.1 KQ971343 KYB27391.1 AXCN02002061 AXCM01002738 AGBW02011079 OWR47243.1 KPJ11382.1 AJVK01059427 EFA03208.2 GFDF01000889 JAV13195.1 APCN01001715 AB047488 BR000546 BAB32485.1 FAA00548.1 GFXV01003593 MBW15398.1 CAQQ02175411 CAQQ02175412 AAZO01007414 NNAY01000425 OXU28499.1 CH954179 KQS62844.1 KK855214 PTY21474.1 GL888812 EGI57987.1 MG601677 AYA50002.1 DS235882 EEB20215.1 AJWK01019933 AJWK01019934 KQ762842 OAD55409.1 CH940647 EDW68490.2 KQ976818 KYN08085.1 OUUW01000009 SPP85132.1 GEBQ01011820 JAT28157.1 KA644545 AFP59174.1 KQ434835 KZC07899.1 KZ288251 PBC31053.1 KQ435696 KOX80821.1 GGMR01018396 MBY31015.1 CH379069 EAL29561.2 KQ436234 KOX67393.1 ADMH02000458 ETN66397.1 CH902618 EDV41228.1 CM000159 EDW93613.1 CM002911 KMY93176.1 CM000158 EDW90785.2 CH933809 EDW17477.2 GL762903 EFZ20536.1 AE013599 ABV53776.1 KQ982482 KYQ55918.1 CVRI01000054 CRK99894.1 CH480816 EDW47551.1 KQ414716 KOC62803.1 CM000362 EDX06752.1 ABLF02030255 ABLF02030257 AK341268 BAH71700.1 GL448255 EFN85030.1 KOC62801.1
KOB70397.1 AGBW02009345 OWR51056.1 RSAL01000156 RVE45634.1 AK401846 BAM18468.1 KQ459586 KPI97913.1 KQ460883 KPJ11369.1 GFTR01002672 JAW13754.1 GAHY01000221 JAA77289.1 ACPB03017958 ACPB03017959 KK855351 PTY22120.1 MF942789 AWK28312.1 GAHY01002330 JAA75180.1 ACPB03009497 LJIG01000626 KRT86314.1 GEMB01000898 JAS02243.1 GDHC01001027 JAQ17602.1 GBHO01020110 JAG23494.1 CH477267 EAT45440.1 PYGN01001077 PSN37817.1 NEVH01006738 PNF36609.1 KPI97900.1 ODYU01001934 SOQ38827.1 NWSH01003794 PCG65814.1 NWSH01007872 PCG62774.1 GBHO01003344 JAG40260.1 AAAB01008980 EAA14379.5 ACPB03017960 GAHY01002338 JAA75172.1 GDHC01017817 JAQ00812.1 KQ971343 KYB27391.1 AXCN02002061 AXCM01002738 AGBW02011079 OWR47243.1 KPJ11382.1 AJVK01059427 EFA03208.2 GFDF01000889 JAV13195.1 APCN01001715 AB047488 BR000546 BAB32485.1 FAA00548.1 GFXV01003593 MBW15398.1 CAQQ02175411 CAQQ02175412 AAZO01007414 NNAY01000425 OXU28499.1 CH954179 KQS62844.1 KK855214 PTY21474.1 GL888812 EGI57987.1 MG601677 AYA50002.1 DS235882 EEB20215.1 AJWK01019933 AJWK01019934 KQ762842 OAD55409.1 CH940647 EDW68490.2 KQ976818 KYN08085.1 OUUW01000009 SPP85132.1 GEBQ01011820 JAT28157.1 KA644545 AFP59174.1 KQ434835 KZC07899.1 KZ288251 PBC31053.1 KQ435696 KOX80821.1 GGMR01018396 MBY31015.1 CH379069 EAL29561.2 KQ436234 KOX67393.1 ADMH02000458 ETN66397.1 CH902618 EDV41228.1 CM000159 EDW93613.1 CM002911 KMY93176.1 CM000158 EDW90785.2 CH933809 EDW17477.2 GL762903 EFZ20536.1 AE013599 ABV53776.1 KQ982482 KYQ55918.1 CVRI01000054 CRK99894.1 CH480816 EDW47551.1 KQ414716 KOC62803.1 CM000362 EDX06752.1 ABLF02030255 ABLF02030257 AK341268 BAH71700.1 GL448255 EFN85030.1 KOC62801.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000015103 UP000008820 UP000245037 UP000235965 UP000075881 UP000218220 UP000075903 UP000075882 UP000076407 UP000007062 UP000075880 UP000007266 UP000075886 UP000075883 UP000092462 UP000075902 UP000075840 UP000075884 UP000075885 UP000075900 UP000005203 UP000015102 UP000215335 UP000002358 UP000075920 UP000192223 UP000008711 UP000007755 UP000009046 UP000092461 UP000076408 UP000008792 UP000192221 UP000078542 UP000268350 UP000069272 UP000095300 UP000095301 UP000076502 UP000242457 UP000053105 UP000001819 UP000000673 UP000007801 UP000002282 UP000009192 UP000000803 UP000075809 UP000183832 UP000001292 UP000053825 UP000000304 UP000007819 UP000008237
UP000015103 UP000008820 UP000245037 UP000235965 UP000075881 UP000218220 UP000075903 UP000075882 UP000076407 UP000007062 UP000075880 UP000007266 UP000075886 UP000075883 UP000092462 UP000075902 UP000075840 UP000075884 UP000075885 UP000075900 UP000005203 UP000015102 UP000215335 UP000002358 UP000075920 UP000192223 UP000008711 UP000007755 UP000009046 UP000092461 UP000076408 UP000008792 UP000192221 UP000078542 UP000268350 UP000069272 UP000095300 UP000095301 UP000076502 UP000242457 UP000053105 UP000001819 UP000000673 UP000007801 UP000002282 UP000009192 UP000000803 UP000075809 UP000183832 UP000001292 UP000053825 UP000000304 UP000007819 UP000008237
ProteinModelPortal
C0H6M5
A0A2H1V805
A0A0L7L4J5
A0A212FBE3
A0A3S2NQ74
I4DKM8
+ More
A0A194PY11 A0A194R1H4 A0A224XRL8 R4G543 T1I0T8 A0A2R7WSK8 A0A2S1ZS73 R4G3C1 T1I4C4 A0A0T6BG89 A0A171ALV7 A0A146MF21 A0A0A9XU59 Q17G30 A0A2P8Y0M8 A0A2J7R701 A0A194Q3B6 A0A2H1VDC4 A0A182KCU6 A0A2A4J2S0 A0A2A4IU20 A0A0A9ZEM1 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 T1I0T7 A0A182IU01 R4FJB9 A0A146L0M8 A0A139WHQ8 A0A182QGA7 A0A182MIF1 A0A212F0I8 A0A194R101 A0A1B0DG98 A0A182TXD1 D6WNV0 A0A1L8E3U5 A0A182HRK7 A0A182NUJ4 A0A182PJH7 A0A182RHG2 A0A088A4T5 Q9BPR1 A0A2H8TQ63 T1GT21 A0A1S4N1V2 A0A232FDH9 K7J4G1 A0A182WGU5 A0A1W4XP64 A0A0Q5VPK8 A0A2R7WPG2 F4X6E7 A0A3Q8HGL4 E0W3K9 A0A1B0GJ98 A0A310SI56 A0A182YDM7 A0A1W4XNY4 B4LHG0 A0A1W4U635 A0A195D5A8 A0A3B0JXJ0 A0A1B6LWW6 A0A1W4XN34 A0A182F693 A0A1I8NPH7 T1P8T8 A0A154P7J2 A0A2A3EGY6 A0A0N1IU57 A0A2S2PNN1 Q2LZE7 A0A0M8ZQ66 W5JPT7 B3MA08 B4PJE5 A0A0J9RAX5 B4P5C3 B4KWN0 E9IFU8 A8DRW0 A0A1W4XP74 A0A151X6J9 A0A1J1ILD8 B4HPA2 A0A0L7QVZ5 B4QCU1 C4WUY0 E2BGZ7 A0A0L7QW49
A0A194PY11 A0A194R1H4 A0A224XRL8 R4G543 T1I0T8 A0A2R7WSK8 A0A2S1ZS73 R4G3C1 T1I4C4 A0A0T6BG89 A0A171ALV7 A0A146MF21 A0A0A9XU59 Q17G30 A0A2P8Y0M8 A0A2J7R701 A0A194Q3B6 A0A2H1VDC4 A0A182KCU6 A0A2A4J2S0 A0A2A4IU20 A0A0A9ZEM1 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 T1I0T7 A0A182IU01 R4FJB9 A0A146L0M8 A0A139WHQ8 A0A182QGA7 A0A182MIF1 A0A212F0I8 A0A194R101 A0A1B0DG98 A0A182TXD1 D6WNV0 A0A1L8E3U5 A0A182HRK7 A0A182NUJ4 A0A182PJH7 A0A182RHG2 A0A088A4T5 Q9BPR1 A0A2H8TQ63 T1GT21 A0A1S4N1V2 A0A232FDH9 K7J4G1 A0A182WGU5 A0A1W4XP64 A0A0Q5VPK8 A0A2R7WPG2 F4X6E7 A0A3Q8HGL4 E0W3K9 A0A1B0GJ98 A0A310SI56 A0A182YDM7 A0A1W4XNY4 B4LHG0 A0A1W4U635 A0A195D5A8 A0A3B0JXJ0 A0A1B6LWW6 A0A1W4XN34 A0A182F693 A0A1I8NPH7 T1P8T8 A0A154P7J2 A0A2A3EGY6 A0A0N1IU57 A0A2S2PNN1 Q2LZE7 A0A0M8ZQ66 W5JPT7 B3MA08 B4PJE5 A0A0J9RAX5 B4P5C3 B4KWN0 E9IFU8 A8DRW0 A0A1W4XP74 A0A151X6J9 A0A1J1ILD8 B4HPA2 A0A0L7QVZ5 B4QCU1 C4WUY0 E2BGZ7 A0A0L7QW49
Ontologies
GO
PANTHER
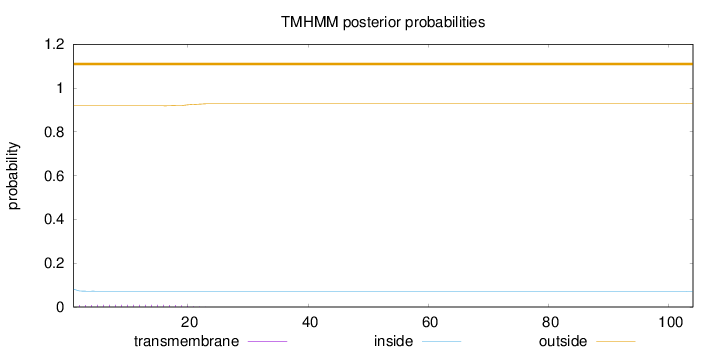
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.997300
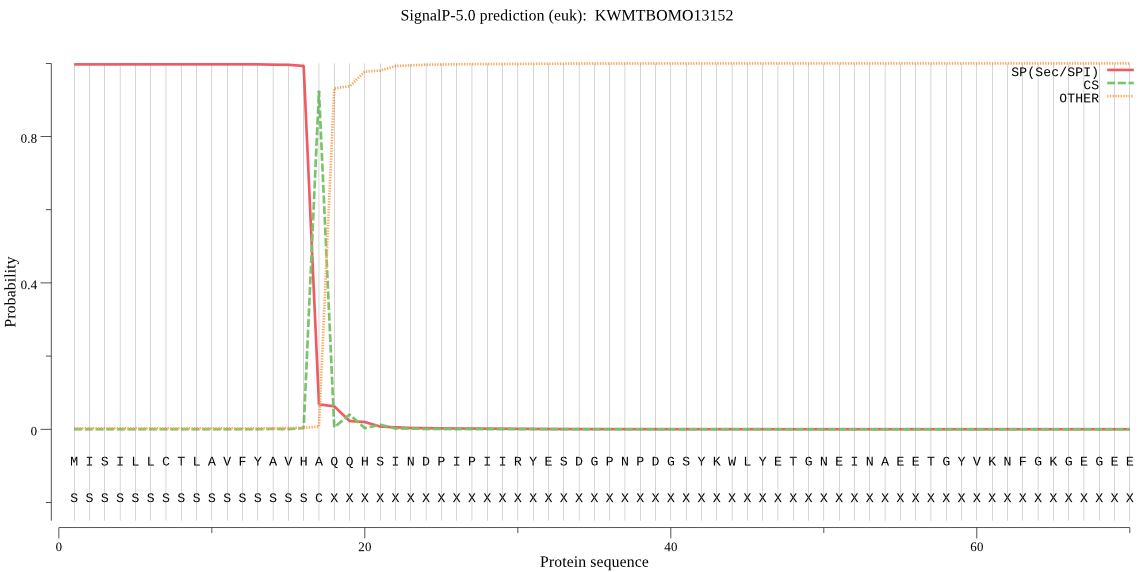
Length:
104
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17533
Exp number, first 60 AAs:
0.17533
Total prob of N-in:
0.08126
outside
1 - 104
Population Genetic Test Statistics
Pi
271.260716
Theta
28.752944
Tajima's D
0.885742
CLR
0.36568
CSRT
0.633418329083546
Interpretation
Uncertain
