Gene
KWMTBOMO13147
Pre Gene Modal
BGIBMGA000334
Annotation
cuticular_protein_RR-1_motif_37_precursor_[Bombyx_mori]
Full name
Endocuticle structural glycoprotein ABD-4
+ More
Endocuticle structural glycoprotein SgAbd-4
Endocuticle structural glycoprotein SgAbd-4
Location in the cell
Cytoplasmic Reliability : 1.347 Extracellular Reliability : 1.713
Sequence
CDS
ATGACGATGAAGTCGATCTTCTTGTTTGGTTTGGTGGCAGCAGCGCTCGCAGCTCCTCAGCCTCAAAAATCAGACCAGACAGCGGAAATCATCAAGCAAGATTTTGATCAGCAAGTTGACGGTTCATACCAGTTCAGCTATGAAACAGACAACGGTATAAAAGCGGAGGAAACGGGCTCTCTGAAAAAGGCAAGCGGCCCGGACGCGAGTGACGTGATCATTGCTCAAGGGGCTTTCAGCTATACAGCACCAGATGGCACCGTCATCTCTCTGAACTACGTTGCTGACGACGACGGTGGCTTCAAGCCTGAGGGTGCTCATTTACCAACGCCGCCGCCGATACCGCCAGCAATCCAGAAGGCCTTAGACTTCCTTGCAACCGCGCCTCCTCCACCATCCTCACGGAACTGA
Protein
MTMKSIFLFGLVAAALAAPQPQKSDQTAEIIKQDFDQQVDGSYQFSYETDNGIKAEETGSLKKASGPDASDVIIAQGAFSYTAPDGTVISLNYVADDDGGFKPEGAHLPTPPPIPPAIQKALDFLATAPPPPSSRN
Summary
Keywords
Amidation
Cuticle
Direct protein sequencing
Glycoprotein
Pyrrolidone carboxylic acid
Feature
chain Endocuticle structural glycoprotein ABD-4
Uniprot
C0H6M9
A0A3S2NA82
A0A212FBD0
A0A194R0Q1
A0A194PYT6
I4DJP2
+ More
A0A182JMG3 B4P5B5 B4NN92 Q17G16 A0A182YDM4 B3NS21 W5JB35 A0A182T7W2 T1E7H0 B4HPB0 B4QCV0 Q95RB2 A0A182V2U4 A0A1W4U6S7 A0A3B0JFU0 A0A182Q5V7 B3MCJ3 A0A182NUJ7 Q290D1 B4GCF1 A0A1A9VUT7 A0A182X789 A0A084W213 A0A182U7B6 A0A182KTN2 Q7Q1B2 A0A182HRL0 A0A0M4EVF5 A0A336M3T7 U5ERP1 A0A182PFJ0 A0A023EGF7 A0A1B0G6Q2 A0A1A9Y1V8 A0A1B0AM99 A0A1B0ABQ2 A0A2H1VNS6 A0A1A9X163 A0A182MSH2 W8BMH3 A0A0A1XCC6 A0A222NTA6 B0WRC2 B4KRV6 A0A034W0D7 A0A0K8VUW7 P21799 B4LLT1 A0A182WGU1 A0A023EDL5 B4J718 A0A336MMY9 A0A0L0CCY2 A0A1I8MQ56 A0A1I8PB46 A0A1Q3FRI9 A0A1Q3FRE6 E0VR57 Q7M4F1 A0A182RHF8 A0A2P8Y5M7 A0A1W4XP74 A0A182F696 A0A023EDG7 D6WNK6 A0A1Y1LMF0 A0A1L8E3R2 A0A224XSV9 T1GT21 A0A161MSD9 R4G2W3 A0A1B0GJ94 B0ZYQ2 A0A3Q8HIK7 B4LLT8 N6TWE4 B4KRW3 B4GCE4 D6WNU5 A0A194Q3B6 A0A182IU01 Q290D7 A0A2H1VDC4 Q17G30 A0A1S4N1V2 B4J726 B3MCI5 A0A222NT99 U4UFY9 A0A0T6BD43 A0A2A4J2S0 A0A2A4IU20
A0A182JMG3 B4P5B5 B4NN92 Q17G16 A0A182YDM4 B3NS21 W5JB35 A0A182T7W2 T1E7H0 B4HPB0 B4QCV0 Q95RB2 A0A182V2U4 A0A1W4U6S7 A0A3B0JFU0 A0A182Q5V7 B3MCJ3 A0A182NUJ7 Q290D1 B4GCF1 A0A1A9VUT7 A0A182X789 A0A084W213 A0A182U7B6 A0A182KTN2 Q7Q1B2 A0A182HRL0 A0A0M4EVF5 A0A336M3T7 U5ERP1 A0A182PFJ0 A0A023EGF7 A0A1B0G6Q2 A0A1A9Y1V8 A0A1B0AM99 A0A1B0ABQ2 A0A2H1VNS6 A0A1A9X163 A0A182MSH2 W8BMH3 A0A0A1XCC6 A0A222NTA6 B0WRC2 B4KRV6 A0A034W0D7 A0A0K8VUW7 P21799 B4LLT1 A0A182WGU1 A0A023EDL5 B4J718 A0A336MMY9 A0A0L0CCY2 A0A1I8MQ56 A0A1I8PB46 A0A1Q3FRI9 A0A1Q3FRE6 E0VR57 Q7M4F1 A0A182RHF8 A0A2P8Y5M7 A0A1W4XP74 A0A182F696 A0A023EDG7 D6WNK6 A0A1Y1LMF0 A0A1L8E3R2 A0A224XSV9 T1GT21 A0A161MSD9 R4G2W3 A0A1B0GJ94 B0ZYQ2 A0A3Q8HIK7 B4LLT8 N6TWE4 B4KRW3 B4GCE4 D6WNU5 A0A194Q3B6 A0A182IU01 Q290D7 A0A2H1VDC4 Q17G30 A0A1S4N1V2 B4J726 B3MCI5 A0A222NT99 U4UFY9 A0A0T6BD43 A0A2A4J2S0 A0A2A4IU20
Pubmed
19121390
19280704
22118469
26354079
22651552
17994087
+ More
17550304 17510324 25244985 20920257 23761445 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 24438588 20966253 12364791 14747013 17210077 24945155 24495485 25830018 25348373 1997327 26108605 25315136 20566863 9692242 29403074 18362917 19820115 28004739 23537049
17550304 17510324 25244985 20920257 23761445 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 24438588 20966253 12364791 14747013 17210077 24945155 24495485 25830018 25348373 1997327 26108605 25315136 20566863 9692242 29403074 18362917 19820115 28004739 23537049
EMBL
BABH01013061
BR000538
FAA00540.1
RSAL01000156
RVE45630.1
AGBW02009345
+ More
OWR51052.1 KQ460883 KPJ11373.1 KQ459586 KPI97909.1 AK401510 BAM18132.1 CM000158 EDW90777.1 CH964282 EDW85831.1 CH477267 EAT45454.1 CH954179 EDV56323.1 ADMH02002020 ETN59974.1 GAMD01003062 JAA98528.1 CH480816 EDW47559.1 CM000362 CM002911 EDX06761.1 KMY93188.1 AE013599 AY061515 KX531749 AAF58518.2 AAL29063.1 ANY27559.1 OUUW01000001 SPP74200.1 AXCN02002063 CH902619 EDV36227.1 CM000071 EAL25431.3 CH479181 EDW32428.1 ATLV01019473 KE525272 KFB44257.1 AAAB01008980 EAA14421.4 APCN01001718 CP012524 ALC41595.1 UFQT01000134 SSX20658.1 GANO01002726 JAB57145.1 GAPW01005869 JAC07729.1 CCAG010018758 CCAG010018759 JXJN01000377 ODYU01003544 SOQ42456.1 AXCM01008802 GAMC01011979 JAB94576.1 GBXI01006179 JAD08113.1 KX503038 ASQ42724.1 DS232054 EDS33312.1 CH933808 EDW09397.1 GAKP01011332 JAC47620.1 GDHF01023862 GDHF01009643 JAI28452.1 JAI42671.1 CH940648 EDW61954.1 GAPW01006010 JAC07588.1 CH916367 EDW01006.1 UFQT01001422 SSX30489.1 JRES01000678 KNC29324.1 GFDL01004868 JAV30177.1 GFDL01004845 JAV30200.1 DS235451 EEB15863.1 PYGN01000899 PSN39556.1 GAPW01006130 JAC07468.1 KQ971343 EFA03215.1 GEZM01057662 JAV72177.1 GFDF01000723 JAV13361.1 GFTR01002232 JAW14194.1 CAQQ02175411 CAQQ02175412 GEMB01000891 JAS02249.1 ACPB03023185 GAHY01002236 JAA75274.1 AJWK01019926 AJWK01019927 EU407178 ABZ04123.1 MG601680 AYA50005.1 EDW61961.1 APGK01052420 KB741212 ENN72716.1 EDW09404.1 EDW32421.1 EFA03211.1 KPI97900.1 EAL25425.3 ODYU01001934 SOQ38827.1 EAT45440.1 AAZO01007414 EDW01014.1 EDV36219.1 KX503041 ASQ42727.1 KB632279 ERL91283.1 LJIG01001715 KRT85225.1 NWSH01003794 PCG65814.1 NWSH01007872 PCG62774.1
OWR51052.1 KQ460883 KPJ11373.1 KQ459586 KPI97909.1 AK401510 BAM18132.1 CM000158 EDW90777.1 CH964282 EDW85831.1 CH477267 EAT45454.1 CH954179 EDV56323.1 ADMH02002020 ETN59974.1 GAMD01003062 JAA98528.1 CH480816 EDW47559.1 CM000362 CM002911 EDX06761.1 KMY93188.1 AE013599 AY061515 KX531749 AAF58518.2 AAL29063.1 ANY27559.1 OUUW01000001 SPP74200.1 AXCN02002063 CH902619 EDV36227.1 CM000071 EAL25431.3 CH479181 EDW32428.1 ATLV01019473 KE525272 KFB44257.1 AAAB01008980 EAA14421.4 APCN01001718 CP012524 ALC41595.1 UFQT01000134 SSX20658.1 GANO01002726 JAB57145.1 GAPW01005869 JAC07729.1 CCAG010018758 CCAG010018759 JXJN01000377 ODYU01003544 SOQ42456.1 AXCM01008802 GAMC01011979 JAB94576.1 GBXI01006179 JAD08113.1 KX503038 ASQ42724.1 DS232054 EDS33312.1 CH933808 EDW09397.1 GAKP01011332 JAC47620.1 GDHF01023862 GDHF01009643 JAI28452.1 JAI42671.1 CH940648 EDW61954.1 GAPW01006010 JAC07588.1 CH916367 EDW01006.1 UFQT01001422 SSX30489.1 JRES01000678 KNC29324.1 GFDL01004868 JAV30177.1 GFDL01004845 JAV30200.1 DS235451 EEB15863.1 PYGN01000899 PSN39556.1 GAPW01006130 JAC07468.1 KQ971343 EFA03215.1 GEZM01057662 JAV72177.1 GFDF01000723 JAV13361.1 GFTR01002232 JAW14194.1 CAQQ02175411 CAQQ02175412 GEMB01000891 JAS02249.1 ACPB03023185 GAHY01002236 JAA75274.1 AJWK01019926 AJWK01019927 EU407178 ABZ04123.1 MG601680 AYA50005.1 EDW61961.1 APGK01052420 KB741212 ENN72716.1 EDW09404.1 EDW32421.1 EFA03211.1 KPI97900.1 EAL25425.3 ODYU01001934 SOQ38827.1 EAT45440.1 AAZO01007414 EDW01014.1 EDV36219.1 KX503041 ASQ42727.1 KB632279 ERL91283.1 LJIG01001715 KRT85225.1 NWSH01003794 PCG65814.1 NWSH01007872 PCG62774.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000075880
+ More
UP000002282 UP000007798 UP000008820 UP000076408 UP000008711 UP000000673 UP000075901 UP000001292 UP000000304 UP000000803 UP000075903 UP000192221 UP000268350 UP000075886 UP000007801 UP000075884 UP000001819 UP000008744 UP000078200 UP000076407 UP000030765 UP000075902 UP000075882 UP000007062 UP000075840 UP000092553 UP000075885 UP000092444 UP000092443 UP000092460 UP000092445 UP000091820 UP000075883 UP000002320 UP000009192 UP000008792 UP000075920 UP000001070 UP000037069 UP000095301 UP000095300 UP000009046 UP000075900 UP000245037 UP000192223 UP000069272 UP000007266 UP000015102 UP000015103 UP000092461 UP000019118 UP000030742 UP000218220
UP000002282 UP000007798 UP000008820 UP000076408 UP000008711 UP000000673 UP000075901 UP000001292 UP000000304 UP000000803 UP000075903 UP000192221 UP000268350 UP000075886 UP000007801 UP000075884 UP000001819 UP000008744 UP000078200 UP000076407 UP000030765 UP000075902 UP000075882 UP000007062 UP000075840 UP000092553 UP000075885 UP000092444 UP000092443 UP000092460 UP000092445 UP000091820 UP000075883 UP000002320 UP000009192 UP000008792 UP000075920 UP000001070 UP000037069 UP000095301 UP000095300 UP000009046 UP000075900 UP000245037 UP000192223 UP000069272 UP000007266 UP000015102 UP000015103 UP000092461 UP000019118 UP000030742 UP000218220
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6M9
A0A3S2NA82
A0A212FBD0
A0A194R0Q1
A0A194PYT6
I4DJP2
+ More
A0A182JMG3 B4P5B5 B4NN92 Q17G16 A0A182YDM4 B3NS21 W5JB35 A0A182T7W2 T1E7H0 B4HPB0 B4QCV0 Q95RB2 A0A182V2U4 A0A1W4U6S7 A0A3B0JFU0 A0A182Q5V7 B3MCJ3 A0A182NUJ7 Q290D1 B4GCF1 A0A1A9VUT7 A0A182X789 A0A084W213 A0A182U7B6 A0A182KTN2 Q7Q1B2 A0A182HRL0 A0A0M4EVF5 A0A336M3T7 U5ERP1 A0A182PFJ0 A0A023EGF7 A0A1B0G6Q2 A0A1A9Y1V8 A0A1B0AM99 A0A1B0ABQ2 A0A2H1VNS6 A0A1A9X163 A0A182MSH2 W8BMH3 A0A0A1XCC6 A0A222NTA6 B0WRC2 B4KRV6 A0A034W0D7 A0A0K8VUW7 P21799 B4LLT1 A0A182WGU1 A0A023EDL5 B4J718 A0A336MMY9 A0A0L0CCY2 A0A1I8MQ56 A0A1I8PB46 A0A1Q3FRI9 A0A1Q3FRE6 E0VR57 Q7M4F1 A0A182RHF8 A0A2P8Y5M7 A0A1W4XP74 A0A182F696 A0A023EDG7 D6WNK6 A0A1Y1LMF0 A0A1L8E3R2 A0A224XSV9 T1GT21 A0A161MSD9 R4G2W3 A0A1B0GJ94 B0ZYQ2 A0A3Q8HIK7 B4LLT8 N6TWE4 B4KRW3 B4GCE4 D6WNU5 A0A194Q3B6 A0A182IU01 Q290D7 A0A2H1VDC4 Q17G30 A0A1S4N1V2 B4J726 B3MCI5 A0A222NT99 U4UFY9 A0A0T6BD43 A0A2A4J2S0 A0A2A4IU20
A0A182JMG3 B4P5B5 B4NN92 Q17G16 A0A182YDM4 B3NS21 W5JB35 A0A182T7W2 T1E7H0 B4HPB0 B4QCV0 Q95RB2 A0A182V2U4 A0A1W4U6S7 A0A3B0JFU0 A0A182Q5V7 B3MCJ3 A0A182NUJ7 Q290D1 B4GCF1 A0A1A9VUT7 A0A182X789 A0A084W213 A0A182U7B6 A0A182KTN2 Q7Q1B2 A0A182HRL0 A0A0M4EVF5 A0A336M3T7 U5ERP1 A0A182PFJ0 A0A023EGF7 A0A1B0G6Q2 A0A1A9Y1V8 A0A1B0AM99 A0A1B0ABQ2 A0A2H1VNS6 A0A1A9X163 A0A182MSH2 W8BMH3 A0A0A1XCC6 A0A222NTA6 B0WRC2 B4KRV6 A0A034W0D7 A0A0K8VUW7 P21799 B4LLT1 A0A182WGU1 A0A023EDL5 B4J718 A0A336MMY9 A0A0L0CCY2 A0A1I8MQ56 A0A1I8PB46 A0A1Q3FRI9 A0A1Q3FRE6 E0VR57 Q7M4F1 A0A182RHF8 A0A2P8Y5M7 A0A1W4XP74 A0A182F696 A0A023EDG7 D6WNK6 A0A1Y1LMF0 A0A1L8E3R2 A0A224XSV9 T1GT21 A0A161MSD9 R4G2W3 A0A1B0GJ94 B0ZYQ2 A0A3Q8HIK7 B4LLT8 N6TWE4 B4KRW3 B4GCE4 D6WNU5 A0A194Q3B6 A0A182IU01 Q290D7 A0A2H1VDC4 Q17G30 A0A1S4N1V2 B4J726 B3MCI5 A0A222NT99 U4UFY9 A0A0T6BD43 A0A2A4J2S0 A0A2A4IU20
Ontologies
GO
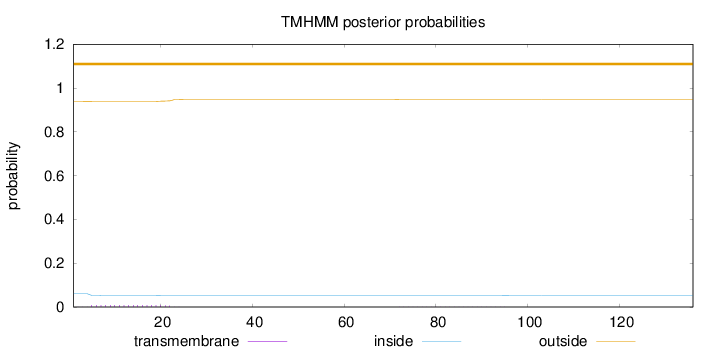
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.990420
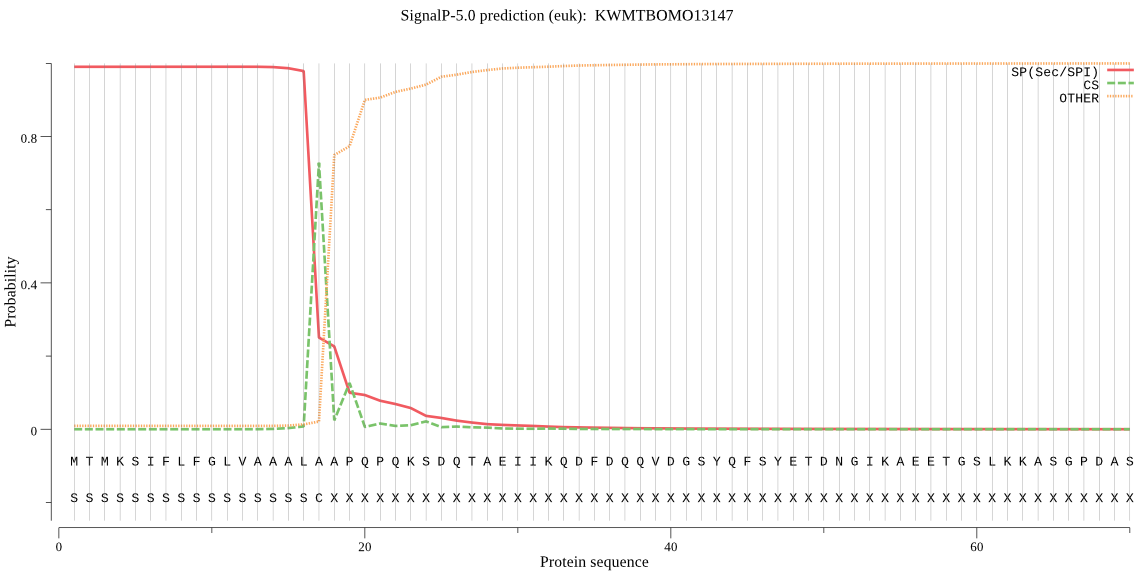
Length:
136
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18215
Exp number, first 60 AAs:
0.15476
Total prob of N-in:
0.06066
outside
1 - 136
Population Genetic Test Statistics
Pi
249.179627
Theta
187.028011
Tajima's D
1.169285
CLR
0.004331
CSRT
0.697865106744663
Interpretation
Uncertain
