Pre Gene Modal
BGIBMGA000332
Annotation
cuticular_protein_RR-1_motif_39_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.742 PlasmaMembrane Reliability : 1.286
Sequence
CDS
ATGAAATTCTTTGTGGTATTCGCAGCTGTTGTAGCTGTTTCGGCTGCTCAGTTTGGAGGATTTATCCAGTCAACTCGGATCCCTGGCTTCCCTCGGCCTACTATACCACCACCTCCACCACCCAGACCGACCATTCAGGCTATTCTTCCTAACCGTGTCGCAACTAATGAGCAGGCTGCTGAAACTATCAGTTATCAGAACGAGATCCTCCCTGACGGCAGCTACAGCCATGGGTTTGAGACTAACAACGGAATCTCAGCTCAAGCACAAGGAACTCCTCGTGACTTCGGAGGCAATCCTCCCGTTGTGCCTGTAGTATCCCAGGGATCGTTCGCCTGGACATCTCCCGAAGGTCAACCAATCGTAATTACCTACATCGCTGACGAGAACGGCTACCAACCTCAGGGTGATGCTATCCCCACTCCACCTCCAATCCCAGAAGCCATCCTCAGGGCTCTGGACTTCATCGCAAGAAACCCACCAGCGCCCCAGAAGAAGTGA
Protein
MKFFVVFAAVVAVSAAQFGGFIQSTRIPGFPRPTIPPPPPPRPTIQAILPNRVATNEQAAETISYQNEILPDGSYSHGFETNNGISAQAQGTPRDFGGNPPVVPVVSQGSFAWTSPEGQPIVITYIADENGYQPQGDAIPTPPPIPEAILRALDFIARNPPAPQKK
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
Ontologies
GO
Topology
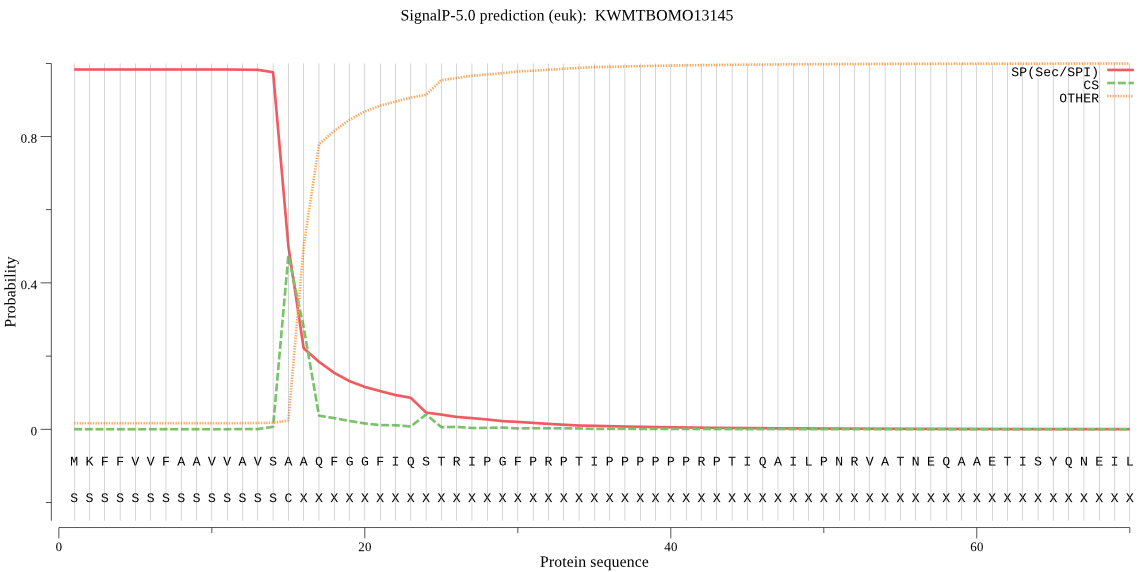
SignalP
Position: 1 - 15,
Likelihood: 0.982082
Length:
166
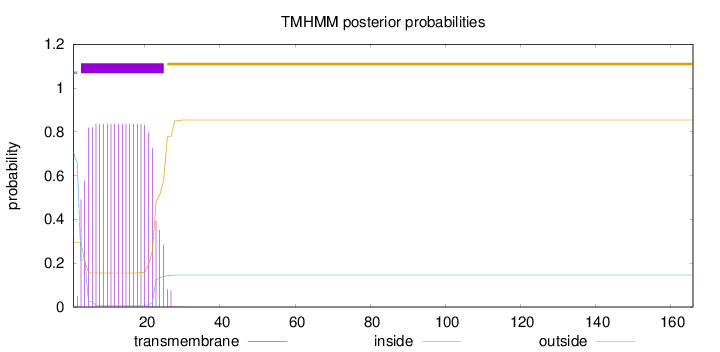
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.18805
Exp number, first 60 AAs:
17.1873
Total prob of N-in:
0.70661
POSSIBLE N-term signal
sequence
inside
1 - 2
TMhelix
3 - 25
outside
26 - 166
Population Genetic Test Statistics
Pi
28.246578
Theta
35.226257
Tajima's D
-0.570334
CLR
0.139012
CSRT
0.222638868056597
Interpretation
Uncertain
