Gene
KWMTBOMO13139
Pre Gene Modal
BGIBMGA000327
Annotation
cuticular_protein_RR-1_motif_43_precursor_[Bombyx_mori]
Full name
Larval cuticle protein LCP-17
Location in the cell
Extracellular Reliability : 2.641
Sequence
CDS
ATGAAGACCTTTGTTATTCTCTTCGGACTTGTGACGGTCGCGGTAGCTGCACCCCAATCACCTACTGAACCCATCCCGATCCTGAAGCAGGAGAGCAGCATCGAGCCTGATGGGTCCTACCAATACTCTTATGAGACTGGAAATGGTATCTCCGCTGCCGAGCGAGGCGCTCTCAAAAACATCGGCGCTGAAGAACCTGCTTTACAGGTTGAAGGTCAATTCCAGTACCCAAGCGAGGACGGTGGTACTATCCAGTTGTCCTACATCGCCAATGAGAACGGCTTCCAGCCTCAGGGATCACATTTACCCACACCACCCCCAATTCCTGAAGTCATCCAGCGCGCTCTCGCATACCTCGCCACCGCACCCCCACAGCCTGAAAACAACAGGCCATAA
Protein
MKTFVILFGLVTVAVAAPQSPTEPIPILKQESSIEPDGSYQYSYETGNGISAAERGALKNIGAEEPALQVEGQFQYPSEDGGTIQLSYIANENGFQPQGSHLPTPPPIPEVIQRALAYLATAPPQPENNRP
Summary
Keywords
Complete proteome
Cuticle
Reference proteome
Signal
Feature
chain Larval cuticle protein LCP-17
Uniprot
C0H6N5
A0A2H1VDB8
G8FVP6
A0A194PX52
A0A194R2A2
A0A212F0G5
+ More
A0A212F0M0 A0A194PX20 A0A194R690 C0H6N6 A0A2H1VDH2 A0A1W4XN34 A0A1W4XNY4 A0A194Q3B6 A0A2A4J2S0 A0A2A4IU20 V5IB19 T1P8T8 F4X6E9 A0A194R101 A0A195DX36 D6WP14 A0A158NJP8 A0A2J7R753 A0A2H1VDC4 A0A1I8NPH7 A0A2H1X3S9 A0A151X6A2 A0A0T6BD43 A0A195BBN7 C0H6M4 A0A212F0I8 Q9BPR1 Q17G30 A0A195FN03 A0A182MFL3 H9J6Y5 A0A182IU01 E9IFV0 W0GHP6 O02387 A0A182IL85 C0H6J3 A0A0T6BG89 A0A2A4J2I4 B2DBG7 A0A182QNQ5 A0A336MJ94 A0A158NFL8 E2B0E6 A0A182QGA7 A0A182S6R8 A0A194Q7B5 A0A2S1ZS73 A0A1J1ILD8 A0A336LRL8 G8FVQ1 A0A0L7QW49 Q290D7 A0A1B0D633 A0A0N1PIW1 A0A182MIF1 A0A182YDM4 D6WNV0 A0A0L7LBG3 A0A194R0P7 A0A182RHG2 F4X6E7 A0A2P8Y5M5 A0A232FDH9 K7J4G1 A0A195D5A8 A0A088A4U8 A0A2A3EJ46 A0A232FDB1 Q7Q1A8 A0A0K8UZ57 A0A182KTN1 A0A0A1WLS1 A0A1J1II08 A0A195D5C7 A0A154P9J5 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 A0A2A4IY21 A0A182U3Y3 A0A182KCU6 B4GCE4 A0A182VEM2 A0A067R439 A0A151J736 A0A1S4N1V2 A0A182TXD1 A0A1B6GX67 A0A1W4XDV2 A0A2S1ZS74 E2BGZ6 E0W3K9
A0A212F0M0 A0A194PX20 A0A194R690 C0H6N6 A0A2H1VDH2 A0A1W4XN34 A0A1W4XNY4 A0A194Q3B6 A0A2A4J2S0 A0A2A4IU20 V5IB19 T1P8T8 F4X6E9 A0A194R101 A0A195DX36 D6WP14 A0A158NJP8 A0A2J7R753 A0A2H1VDC4 A0A1I8NPH7 A0A2H1X3S9 A0A151X6A2 A0A0T6BD43 A0A195BBN7 C0H6M4 A0A212F0I8 Q9BPR1 Q17G30 A0A195FN03 A0A182MFL3 H9J6Y5 A0A182IU01 E9IFV0 W0GHP6 O02387 A0A182IL85 C0H6J3 A0A0T6BG89 A0A2A4J2I4 B2DBG7 A0A182QNQ5 A0A336MJ94 A0A158NFL8 E2B0E6 A0A182QGA7 A0A182S6R8 A0A194Q7B5 A0A2S1ZS73 A0A1J1ILD8 A0A336LRL8 G8FVQ1 A0A0L7QW49 Q290D7 A0A1B0D633 A0A0N1PIW1 A0A182MIF1 A0A182YDM4 D6WNV0 A0A0L7LBG3 A0A194R0P7 A0A182RHG2 F4X6E7 A0A2P8Y5M5 A0A232FDH9 K7J4G1 A0A195D5A8 A0A088A4U8 A0A2A3EJ46 A0A232FDB1 Q7Q1A8 A0A0K8UZ57 A0A182KTN1 A0A0A1WLS1 A0A1J1II08 A0A195D5C7 A0A154P9J5 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 A0A2A4IY21 A0A182U3Y3 A0A182KCU6 B4GCE4 A0A182VEM2 A0A067R439 A0A151J736 A0A1S4N1V2 A0A182TXD1 A0A1B6GX67 A0A1W4XDV2 A0A2S1ZS74 E2BGZ6 E0W3K9
Pubmed
EMBL
BABH01013076
BR000544
FAA00546.1
ODYU01001934
SOQ38825.1
JN082716
+ More
AER27814.1 KQ459586 KPI97902.1 KQ460883 KPJ11380.1 AGBW02011079 OWR47245.1 OWR47244.1 KPI97901.1 KPJ11381.1 BR000545 FAA00547.1 SOQ38826.1 KPI97900.1 NWSH01003794 PCG65814.1 NWSH01007872 PCG62774.1 GALX01000205 JAB68261.1 KA644545 AFP59174.1 GL888812 EGI57989.1 KPJ11382.1 KQ980155 KYN17460.1 KQ971343 EFA04420.1 ADTU01018177 NEVH01006738 PNF36675.1 SOQ38827.1 ODYU01013214 SOQ59908.1 KQ982482 KYQ55916.1 LJIG01001715 KRT85225.1 KQ976526 KYM81953.1 BABH01013051 BABH01013052 BR000533 FAA00535.1 OWR47243.1 AB047488 BR000546 BAB32485.1 FAA00548.1 CH477267 EAT45440.1 KQ981424 KYN41853.1 AXCM01002738 BABH01028447 GL762903 EFZ20561.1 KF672849 AHF55751.1 AB004766 BR000503 FAA00504.1 LJIG01000626 KRT86314.1 NWSH01003986 PCG65593.1 AB264646 BAG30726.1 AXCN02002061 UFQT01001422 SSX30494.1 ADTU01014660 GL444552 EFN60841.1 KQ459580 KPI99295.1 MF942789 AWK28312.1 CVRI01000054 CRK99894.1 UFQT01000134 SSX20662.1 JN082721 AER27818.1 KQ414716 KOC62801.1 CM000071 EAL25425.3 AJVK01025685 KQ460571 KPJ13960.1 EFA03208.2 JTDY01001912 KOB72556.1 KPJ11368.1 EGI57987.1 PYGN01000899 PSN39557.1 NNAY01000425 OXU28499.1 KQ976818 KYN08085.1 KZ288251 PBC31051.1 OXU28500.1 AAAB01008980 EAA14443.4 GDHF01020357 JAI31957.1 GBXI01014661 JAC99630.1 CRK99895.1 KYN08082.1 KQ434835 KZC07898.1 EAA14379.5 NWSH01005682 PCG64060.1 CH479181 EDW32421.1 KK852994 KDR12717.1 KQ979748 KYN19356.1 AAZO01007414 GECZ01002760 JAS67009.1 MF942790 AWK28313.1 GL448255 EFN85029.1 DS235882 EEB20215.1
AER27814.1 KQ459586 KPI97902.1 KQ460883 KPJ11380.1 AGBW02011079 OWR47245.1 OWR47244.1 KPI97901.1 KPJ11381.1 BR000545 FAA00547.1 SOQ38826.1 KPI97900.1 NWSH01003794 PCG65814.1 NWSH01007872 PCG62774.1 GALX01000205 JAB68261.1 KA644545 AFP59174.1 GL888812 EGI57989.1 KPJ11382.1 KQ980155 KYN17460.1 KQ971343 EFA04420.1 ADTU01018177 NEVH01006738 PNF36675.1 SOQ38827.1 ODYU01013214 SOQ59908.1 KQ982482 KYQ55916.1 LJIG01001715 KRT85225.1 KQ976526 KYM81953.1 BABH01013051 BABH01013052 BR000533 FAA00535.1 OWR47243.1 AB047488 BR000546 BAB32485.1 FAA00548.1 CH477267 EAT45440.1 KQ981424 KYN41853.1 AXCM01002738 BABH01028447 GL762903 EFZ20561.1 KF672849 AHF55751.1 AB004766 BR000503 FAA00504.1 LJIG01000626 KRT86314.1 NWSH01003986 PCG65593.1 AB264646 BAG30726.1 AXCN02002061 UFQT01001422 SSX30494.1 ADTU01014660 GL444552 EFN60841.1 KQ459580 KPI99295.1 MF942789 AWK28312.1 CVRI01000054 CRK99894.1 UFQT01000134 SSX20662.1 JN082721 AER27818.1 KQ414716 KOC62801.1 CM000071 EAL25425.3 AJVK01025685 KQ460571 KPJ13960.1 EFA03208.2 JTDY01001912 KOB72556.1 KPJ11368.1 EGI57987.1 PYGN01000899 PSN39557.1 NNAY01000425 OXU28499.1 KQ976818 KYN08085.1 KZ288251 PBC31051.1 OXU28500.1 AAAB01008980 EAA14443.4 GDHF01020357 JAI31957.1 GBXI01014661 JAC99630.1 CRK99895.1 KYN08082.1 KQ434835 KZC07898.1 EAA14379.5 NWSH01005682 PCG64060.1 CH479181 EDW32421.1 KK852994 KDR12717.1 KQ979748 KYN19356.1 AAZO01007414 GECZ01002760 JAS67009.1 MF942790 AWK28313.1 GL448255 EFN85029.1 DS235882 EEB20215.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000192223
UP000218220
+ More
UP000095301 UP000007755 UP000078492 UP000007266 UP000005205 UP000235965 UP000095300 UP000075809 UP000078540 UP000008820 UP000078541 UP000075883 UP000075880 UP000075886 UP000000311 UP000075901 UP000183832 UP000053825 UP000001819 UP000092462 UP000076408 UP000037510 UP000075900 UP000245037 UP000215335 UP000002358 UP000078542 UP000005203 UP000242457 UP000007062 UP000075882 UP000076502 UP000075903 UP000076407 UP000075902 UP000075881 UP000008744 UP000027135 UP000008237 UP000009046
UP000095301 UP000007755 UP000078492 UP000007266 UP000005205 UP000235965 UP000095300 UP000075809 UP000078540 UP000008820 UP000078541 UP000075883 UP000075880 UP000075886 UP000000311 UP000075901 UP000183832 UP000053825 UP000001819 UP000092462 UP000076408 UP000037510 UP000075900 UP000245037 UP000215335 UP000002358 UP000078542 UP000005203 UP000242457 UP000007062 UP000075882 UP000076502 UP000075903 UP000076407 UP000075902 UP000075881 UP000008744 UP000027135 UP000008237 UP000009046
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6N5
A0A2H1VDB8
G8FVP6
A0A194PX52
A0A194R2A2
A0A212F0G5
+ More
A0A212F0M0 A0A194PX20 A0A194R690 C0H6N6 A0A2H1VDH2 A0A1W4XN34 A0A1W4XNY4 A0A194Q3B6 A0A2A4J2S0 A0A2A4IU20 V5IB19 T1P8T8 F4X6E9 A0A194R101 A0A195DX36 D6WP14 A0A158NJP8 A0A2J7R753 A0A2H1VDC4 A0A1I8NPH7 A0A2H1X3S9 A0A151X6A2 A0A0T6BD43 A0A195BBN7 C0H6M4 A0A212F0I8 Q9BPR1 Q17G30 A0A195FN03 A0A182MFL3 H9J6Y5 A0A182IU01 E9IFV0 W0GHP6 O02387 A0A182IL85 C0H6J3 A0A0T6BG89 A0A2A4J2I4 B2DBG7 A0A182QNQ5 A0A336MJ94 A0A158NFL8 E2B0E6 A0A182QGA7 A0A182S6R8 A0A194Q7B5 A0A2S1ZS73 A0A1J1ILD8 A0A336LRL8 G8FVQ1 A0A0L7QW49 Q290D7 A0A1B0D633 A0A0N1PIW1 A0A182MIF1 A0A182YDM4 D6WNV0 A0A0L7LBG3 A0A194R0P7 A0A182RHG2 F4X6E7 A0A2P8Y5M5 A0A232FDH9 K7J4G1 A0A195D5A8 A0A088A4U8 A0A2A3EJ46 A0A232FDB1 Q7Q1A8 A0A0K8UZ57 A0A182KTN1 A0A0A1WLS1 A0A1J1II08 A0A195D5C7 A0A154P9J5 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 A0A2A4IY21 A0A182U3Y3 A0A182KCU6 B4GCE4 A0A182VEM2 A0A067R439 A0A151J736 A0A1S4N1V2 A0A182TXD1 A0A1B6GX67 A0A1W4XDV2 A0A2S1ZS74 E2BGZ6 E0W3K9
A0A212F0M0 A0A194PX20 A0A194R690 C0H6N6 A0A2H1VDH2 A0A1W4XN34 A0A1W4XNY4 A0A194Q3B6 A0A2A4J2S0 A0A2A4IU20 V5IB19 T1P8T8 F4X6E9 A0A194R101 A0A195DX36 D6WP14 A0A158NJP8 A0A2J7R753 A0A2H1VDC4 A0A1I8NPH7 A0A2H1X3S9 A0A151X6A2 A0A0T6BD43 A0A195BBN7 C0H6M4 A0A212F0I8 Q9BPR1 Q17G30 A0A195FN03 A0A182MFL3 H9J6Y5 A0A182IU01 E9IFV0 W0GHP6 O02387 A0A182IL85 C0H6J3 A0A0T6BG89 A0A2A4J2I4 B2DBG7 A0A182QNQ5 A0A336MJ94 A0A158NFL8 E2B0E6 A0A182QGA7 A0A182S6R8 A0A194Q7B5 A0A2S1ZS73 A0A1J1ILD8 A0A336LRL8 G8FVQ1 A0A0L7QW49 Q290D7 A0A1B0D633 A0A0N1PIW1 A0A182MIF1 A0A182YDM4 D6WNV0 A0A0L7LBG3 A0A194R0P7 A0A182RHG2 F4X6E7 A0A2P8Y5M5 A0A232FDH9 K7J4G1 A0A195D5A8 A0A088A4U8 A0A2A3EJ46 A0A232FDB1 Q7Q1A8 A0A0K8UZ57 A0A182KTN1 A0A0A1WLS1 A0A1J1II08 A0A195D5C7 A0A154P9J5 A0A182UZ29 A0A182KTN8 A0A182X793 Q7PM44 A0A2A4IY21 A0A182U3Y3 A0A182KCU6 B4GCE4 A0A182VEM2 A0A067R439 A0A151J736 A0A1S4N1V2 A0A182TXD1 A0A1B6GX67 A0A1W4XDV2 A0A2S1ZS74 E2BGZ6 E0W3K9
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.997800
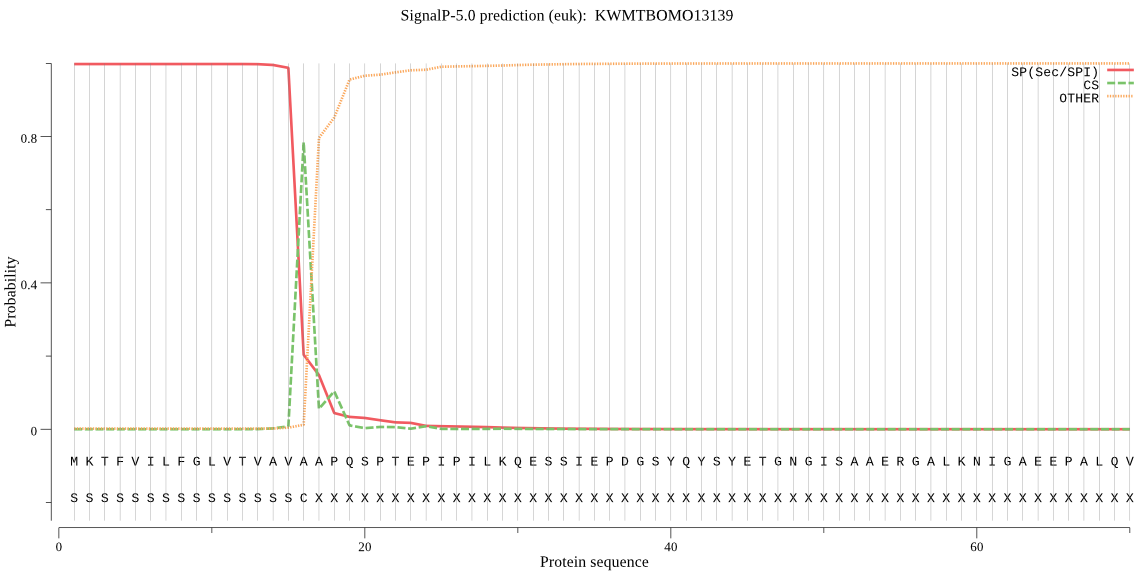
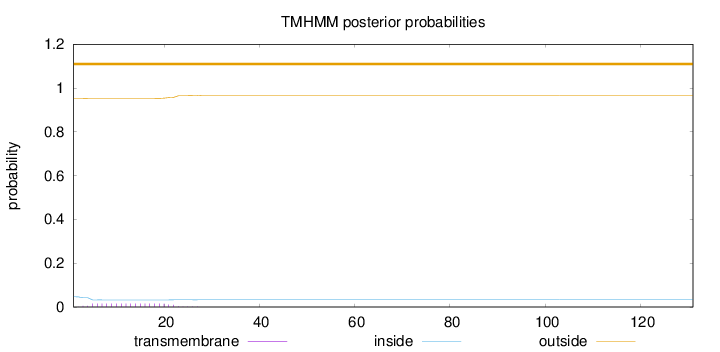
Length:
131
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28959
Exp number, first 60 AAs:
0.28959
Total prob of N-in:
0.04704
outside
1 - 131
Population Genetic Test Statistics
Pi
207.870231
Theta
168.146589
Tajima's D
0.743063
CLR
30.904674
CSRT
0.586220688965552
Interpretation
Uncertain
