Gene
KWMTBOMO13138
Pre Gene Modal
BGIBMGA000326
Annotation
cuticular_protein_RR-1_motif_44_precursor_[Bombyx_mori]
Full name
Endocuticle structural glycoprotein SgAbd-2
+ More
Endocuticle structural glycoprotein SgAbd-8
Endocuticle structural glycoprotein SgAbd-8
Location in the cell
Extracellular Reliability : 2.188
Sequence
CDS
ATGAAATCTATTGTGTTGGTTTCTATGCTGGTCGCAGTGGCCTTGGCCGCGCCTCAGGGGCCCACCGAGCCTATCCCAATTGTTCGTGACGACAGCCAAATCAACGGCGACGGGTCATACCAGTACGCTTTCGAAACCGGCAACGGTATTAGCGCTGACCAGAAGGGTGAACTGAAGAAGGTTGGCGACGTTGAGGCTCTCGAAGTTCAGGGCGAATTCAAATATCCCGGAGAAAACGGTCAGGACATCTCTCTGACCTACACGGCTGATGAGAATGGCTTCCATCCATCCGGCAGCCACTTGCCAACCAGTCCCCCAATCCCGGAAGCCATCCAGCGCGCTCTCGACTTCATCGCCACCGCCCCTCCTGCCCCTGCAGCCGCTGCCGCACAATAG
Protein
MKSIVLVSMLVAVALAAPQGPTEPIPIVRDDSQINGDGSYQYAFETGNGISADQKGELKKVGDVEALEVQGEFKYPGENGQDISLTYTADENGFHPSGSHLPTSPPIPEAIQRALDFIATAPPAPAAAAAQ
Summary
Keywords
Cuticle
Direct protein sequencing
Glycoprotein
Pyrrolidone carboxylic acid
Feature
chain Endocuticle structural glycoprotein SgAbd-2
Uniprot
C0H6N6
A0A194PX20
A0A2H1VDH2
A0A194R690
A0A212F0M0
A0A194PX52
+ More
A0A194R2A2 A0A2H1VDB8 C0H6N5 A0A212F0G5 G8FVP6 A0A1W4XNY4 A0A1W4XN34 A0A2J7R753 A0A182QNQ5 A0A182IL85 D6WFI5 A0A2A4J2I4 A0A1W4XP69 A0A182S6R8 A0A2H1X3S9 Q9BPR1 V5IB19 A0A182MFL3 A0A232FDH9 K7J4G1 G8FVQ1 A0A194Q3B6 A0A1I8NPH7 A0A2A4J2S0 A0A2A4IU20 A0A2H1VDC4 A0A0T6BD43 A0A2H1W3T6 A0A182RHG2 Q7Q1A8 C0H6M4 A0A182KTN1 A0A182U3Y3 A0A1S4N1V2 A0A194R0P7 A0A182VEM2 A0A182KCU6 E0W3K9 Q17G29 A0A212F0I8 T1P8T8 D6WFI1 D6WFI6 D6WFI2 A0A1W4XDV2 A0A194R101 A0A1S4F436 D6WNV0 E9IFU9 A0A2P8XYP8 A0A1B0DG98 A0A3S2N9Y2 A0A2H1W3Z0 Q7M4F3 B0WRB7 A0A2J7R6Z4 I4DIE4 B4LHG0 F4X6E7 A0A182F693 G8FVP9 A0A182WGU5 A0A195DXL6 A0A2P8Y0M8 A0A222NT99 C0H6J9 H9JBX0 Q7M4F2 A0A182X792 A0A2R7WSK8 A0A182MIF1 R4G3X3 A0A0L7QW49 A0A1S3DGD9 A0A1J1ILD8 A0A1L8E3U5 A0A158NJP7 A0A195BCK5 A0A2A3EJ46 E2BGZ6 A0A182IU01 A0A1L8E3R2 Q17G30 A0A1B6GX67 A0A151X6J9 A0A2J7R701 A0A182YDM7 A0A182HRK7 A0A195D5B9 A0A1W4XP64 N6TWE4
A0A194R2A2 A0A2H1VDB8 C0H6N5 A0A212F0G5 G8FVP6 A0A1W4XNY4 A0A1W4XN34 A0A2J7R753 A0A182QNQ5 A0A182IL85 D6WFI5 A0A2A4J2I4 A0A1W4XP69 A0A182S6R8 A0A2H1X3S9 Q9BPR1 V5IB19 A0A182MFL3 A0A232FDH9 K7J4G1 G8FVQ1 A0A194Q3B6 A0A1I8NPH7 A0A2A4J2S0 A0A2A4IU20 A0A2H1VDC4 A0A0T6BD43 A0A2H1W3T6 A0A182RHG2 Q7Q1A8 C0H6M4 A0A182KTN1 A0A182U3Y3 A0A1S4N1V2 A0A194R0P7 A0A182VEM2 A0A182KCU6 E0W3K9 Q17G29 A0A212F0I8 T1P8T8 D6WFI1 D6WFI6 D6WFI2 A0A1W4XDV2 A0A194R101 A0A1S4F436 D6WNV0 E9IFU9 A0A2P8XYP8 A0A1B0DG98 A0A3S2N9Y2 A0A2H1W3Z0 Q7M4F3 B0WRB7 A0A2J7R6Z4 I4DIE4 B4LHG0 F4X6E7 A0A182F693 G8FVP9 A0A182WGU5 A0A195DXL6 A0A2P8Y0M8 A0A222NT99 C0H6J9 H9JBX0 Q7M4F2 A0A182X792 A0A2R7WSK8 A0A182MIF1 R4G3X3 A0A0L7QW49 A0A1S3DGD9 A0A1J1ILD8 A0A1L8E3U5 A0A158NJP7 A0A195BCK5 A0A2A3EJ46 E2BGZ6 A0A182IU01 A0A1L8E3R2 Q17G30 A0A1B6GX67 A0A151X6J9 A0A2J7R701 A0A182YDM7 A0A182HRK7 A0A195D5B9 A0A1W4XP64 N6TWE4
Pubmed
EMBL
BABH01013076
BR000545
FAA00547.1
KQ459586
KPI97901.1
ODYU01001934
+ More
SOQ38826.1 KQ460883 KPJ11381.1 AGBW02011079 OWR47244.1 KPI97902.1 KPJ11380.1 SOQ38825.1 BR000544 FAA00546.1 OWR47245.1 JN082716 AER27814.1 NEVH01006738 PNF36675.1 AXCN02002061 KQ971327 EFA00928.1 NWSH01003986 PCG65593.1 ODYU01013214 SOQ59908.1 AB047488 BR000546 BAB32485.1 FAA00548.1 GALX01000205 JAB68261.1 AXCM01002738 NNAY01000425 OXU28499.1 JN082721 AER27818.1 KPI97900.1 NWSH01003794 PCG65814.1 NWSH01007872 PCG62774.1 SOQ38827.1 LJIG01001715 KRT85225.1 ODYU01006154 SOQ47759.1 AAAB01008980 EAA14443.4 BABH01013051 BABH01013052 BR000533 FAA00535.1 AAZO01007414 KPJ11368.1 DS235882 EEB20215.1 CH477267 EAT45441.1 OWR47243.1 KA644545 AFP59174.1 EFA00924.1 EFA00929.1 EFA00925.1 KPJ11382.1 KQ971343 EFA03208.2 GL762903 EFZ20555.1 PYGN01001152 PSN37138.1 AJVK01059427 RSAL01000374 RVE42117.1 SOQ47758.1 DS232054 EDS33307.1 PNF36608.1 AK401062 BAM17684.1 KPI97914.1 CH940647 EDW68490.2 GL888812 EGI57987.1 JN082719 AER27816.1 KQ980155 KYN17457.1 PYGN01001077 PSN37817.1 KX503041 ASQ42727.1 BR000509 FAA00510.1 BABH01014839 KK855351 PTY22120.1 GAHY01001845 JAA75665.1 KQ414716 KOC62801.1 CVRI01000054 CRK99894.1 GFDF01000889 JAV13195.1 ADTU01018174 KQ976526 KYM81952.1 KZ288251 PBC31051.1 GL448255 EFN85029.1 GFDF01000723 JAV13361.1 EAT45440.1 GECZ01002760 JAS67009.1 KQ982482 KYQ55918.1 PNF36609.1 APCN01001715 KQ976818 KYN08083.1 APGK01052420 KB741212 ENN72716.1
SOQ38826.1 KQ460883 KPJ11381.1 AGBW02011079 OWR47244.1 KPI97902.1 KPJ11380.1 SOQ38825.1 BR000544 FAA00546.1 OWR47245.1 JN082716 AER27814.1 NEVH01006738 PNF36675.1 AXCN02002061 KQ971327 EFA00928.1 NWSH01003986 PCG65593.1 ODYU01013214 SOQ59908.1 AB047488 BR000546 BAB32485.1 FAA00548.1 GALX01000205 JAB68261.1 AXCM01002738 NNAY01000425 OXU28499.1 JN082721 AER27818.1 KPI97900.1 NWSH01003794 PCG65814.1 NWSH01007872 PCG62774.1 SOQ38827.1 LJIG01001715 KRT85225.1 ODYU01006154 SOQ47759.1 AAAB01008980 EAA14443.4 BABH01013051 BABH01013052 BR000533 FAA00535.1 AAZO01007414 KPJ11368.1 DS235882 EEB20215.1 CH477267 EAT45441.1 OWR47243.1 KA644545 AFP59174.1 EFA00924.1 EFA00929.1 EFA00925.1 KPJ11382.1 KQ971343 EFA03208.2 GL762903 EFZ20555.1 PYGN01001152 PSN37138.1 AJVK01059427 RSAL01000374 RVE42117.1 SOQ47758.1 DS232054 EDS33307.1 PNF36608.1 AK401062 BAM17684.1 KPI97914.1 CH940647 EDW68490.2 GL888812 EGI57987.1 JN082719 AER27816.1 KQ980155 KYN17457.1 PYGN01001077 PSN37817.1 KX503041 ASQ42727.1 BR000509 FAA00510.1 BABH01014839 KK855351 PTY22120.1 GAHY01001845 JAA75665.1 KQ414716 KOC62801.1 CVRI01000054 CRK99894.1 GFDF01000889 JAV13195.1 ADTU01018174 KQ976526 KYM81952.1 KZ288251 PBC31051.1 GL448255 EFN85029.1 GFDF01000723 JAV13361.1 EAT45440.1 GECZ01002760 JAS67009.1 KQ982482 KYQ55918.1 PNF36609.1 APCN01001715 KQ976818 KYN08083.1 APGK01052420 KB741212 ENN72716.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000192223
UP000235965
+ More
UP000075886 UP000075880 UP000007266 UP000218220 UP000075901 UP000075883 UP000215335 UP000002358 UP000095300 UP000075900 UP000007062 UP000075882 UP000075902 UP000075903 UP000075881 UP000009046 UP000008820 UP000095301 UP000245037 UP000092462 UP000283053 UP000002320 UP000008792 UP000007755 UP000069272 UP000075920 UP000078492 UP000076407 UP000053825 UP000079169 UP000183832 UP000005205 UP000078540 UP000242457 UP000008237 UP000075809 UP000076408 UP000075840 UP000078542 UP000019118
UP000075886 UP000075880 UP000007266 UP000218220 UP000075901 UP000075883 UP000215335 UP000002358 UP000095300 UP000075900 UP000007062 UP000075882 UP000075902 UP000075903 UP000075881 UP000009046 UP000008820 UP000095301 UP000245037 UP000092462 UP000283053 UP000002320 UP000008792 UP000007755 UP000069272 UP000075920 UP000078492 UP000076407 UP000053825 UP000079169 UP000183832 UP000005205 UP000078540 UP000242457 UP000008237 UP000075809 UP000076408 UP000075840 UP000078542 UP000019118
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6N6
A0A194PX20
A0A2H1VDH2
A0A194R690
A0A212F0M0
A0A194PX52
+ More
A0A194R2A2 A0A2H1VDB8 C0H6N5 A0A212F0G5 G8FVP6 A0A1W4XNY4 A0A1W4XN34 A0A2J7R753 A0A182QNQ5 A0A182IL85 D6WFI5 A0A2A4J2I4 A0A1W4XP69 A0A182S6R8 A0A2H1X3S9 Q9BPR1 V5IB19 A0A182MFL3 A0A232FDH9 K7J4G1 G8FVQ1 A0A194Q3B6 A0A1I8NPH7 A0A2A4J2S0 A0A2A4IU20 A0A2H1VDC4 A0A0T6BD43 A0A2H1W3T6 A0A182RHG2 Q7Q1A8 C0H6M4 A0A182KTN1 A0A182U3Y3 A0A1S4N1V2 A0A194R0P7 A0A182VEM2 A0A182KCU6 E0W3K9 Q17G29 A0A212F0I8 T1P8T8 D6WFI1 D6WFI6 D6WFI2 A0A1W4XDV2 A0A194R101 A0A1S4F436 D6WNV0 E9IFU9 A0A2P8XYP8 A0A1B0DG98 A0A3S2N9Y2 A0A2H1W3Z0 Q7M4F3 B0WRB7 A0A2J7R6Z4 I4DIE4 B4LHG0 F4X6E7 A0A182F693 G8FVP9 A0A182WGU5 A0A195DXL6 A0A2P8Y0M8 A0A222NT99 C0H6J9 H9JBX0 Q7M4F2 A0A182X792 A0A2R7WSK8 A0A182MIF1 R4G3X3 A0A0L7QW49 A0A1S3DGD9 A0A1J1ILD8 A0A1L8E3U5 A0A158NJP7 A0A195BCK5 A0A2A3EJ46 E2BGZ6 A0A182IU01 A0A1L8E3R2 Q17G30 A0A1B6GX67 A0A151X6J9 A0A2J7R701 A0A182YDM7 A0A182HRK7 A0A195D5B9 A0A1W4XP64 N6TWE4
A0A194R2A2 A0A2H1VDB8 C0H6N5 A0A212F0G5 G8FVP6 A0A1W4XNY4 A0A1W4XN34 A0A2J7R753 A0A182QNQ5 A0A182IL85 D6WFI5 A0A2A4J2I4 A0A1W4XP69 A0A182S6R8 A0A2H1X3S9 Q9BPR1 V5IB19 A0A182MFL3 A0A232FDH9 K7J4G1 G8FVQ1 A0A194Q3B6 A0A1I8NPH7 A0A2A4J2S0 A0A2A4IU20 A0A2H1VDC4 A0A0T6BD43 A0A2H1W3T6 A0A182RHG2 Q7Q1A8 C0H6M4 A0A182KTN1 A0A182U3Y3 A0A1S4N1V2 A0A194R0P7 A0A182VEM2 A0A182KCU6 E0W3K9 Q17G29 A0A212F0I8 T1P8T8 D6WFI1 D6WFI6 D6WFI2 A0A1W4XDV2 A0A194R101 A0A1S4F436 D6WNV0 E9IFU9 A0A2P8XYP8 A0A1B0DG98 A0A3S2N9Y2 A0A2H1W3Z0 Q7M4F3 B0WRB7 A0A2J7R6Z4 I4DIE4 B4LHG0 F4X6E7 A0A182F693 G8FVP9 A0A182WGU5 A0A195DXL6 A0A2P8Y0M8 A0A222NT99 C0H6J9 H9JBX0 Q7M4F2 A0A182X792 A0A2R7WSK8 A0A182MIF1 R4G3X3 A0A0L7QW49 A0A1S3DGD9 A0A1J1ILD8 A0A1L8E3U5 A0A158NJP7 A0A195BCK5 A0A2A3EJ46 E2BGZ6 A0A182IU01 A0A1L8E3R2 Q17G30 A0A1B6GX67 A0A151X6J9 A0A2J7R701 A0A182YDM7 A0A182HRK7 A0A195D5B9 A0A1W4XP64 N6TWE4
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.998699
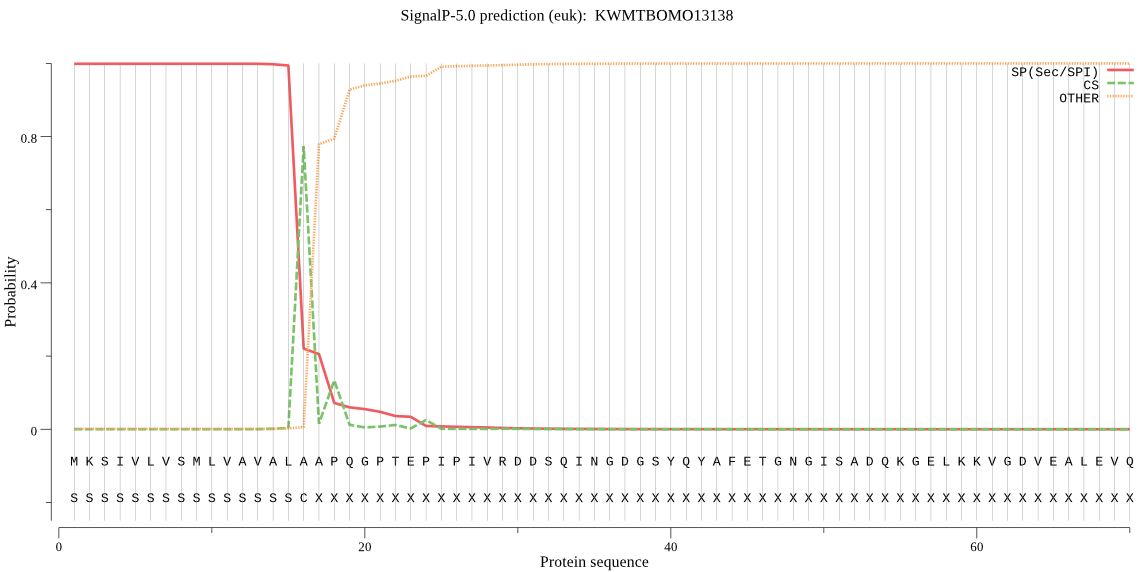
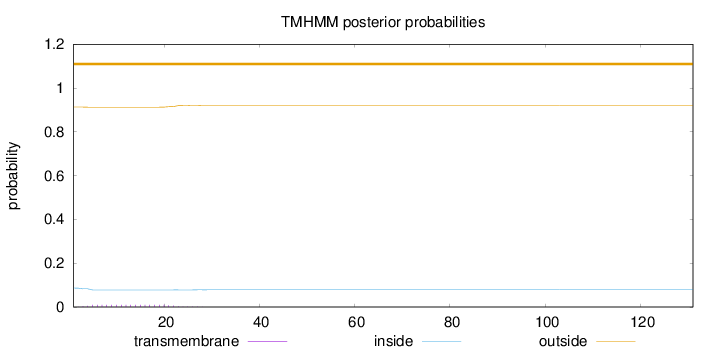
Length:
131
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19718
Exp number, first 60 AAs:
0.19697
Total prob of N-in:
0.08686
outside
1 - 131
Population Genetic Test Statistics
Pi
375.129352
Theta
243.342009
Tajima's D
1.687667
CLR
0
CSRT
0.823958802059897
Interpretation
Uncertain
