Gene
KWMTBOMO13135 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000324
Annotation
cuticular_protein_RR-1_motif_46_precursor_[Bombyx_mori]
Full name
Larval cuticle protein 16/17
+ More
Larval cuticle protein 1
Larval cuticle protein 1
Location in the cell
Extracellular Reliability : 2.792
Sequence
CDS
ATGAAATTGATCATCGTTGTAGCTCTCGCCCTTGTGGCTACCGTCATCGCTGCCCCCCCTACAGAGGACTACCCTAAGATCGTTCGGTCTGAATTCGACGCCAGCCCCGATGGAGCTTACAACTACAATTTCGAGACTTCCAACGGCATCGTGCGTAGCGAAACTGGTGAGCTGAAGGAGGCTCTCGACGACGACAACAAGCCTCACGTTATTGTTGCTGTGCGTGGAAGCTACAGTTACACGAACACTGACGGCAAACCTGAAACCATTACGTACTTCGCTGACGAGACTGGATACCATGCTCAGGGTGAATCTATTCCTCAGGTCGCCTCCGCCACGAGCTAA
Protein
MKLIIVVALALVATVIAAPPTEDYPKIVRSEFDASPDGAYNYNFETSNGIVRSETGELKEALDDDNKPHVIVAVRGSYSYTNTDGKPETITYFADETGYHAQGESIPQVASATS
Summary
Keywords
Cuticle
Polymorphism
Signal
Feature
chain Larval cuticle protein 16/17
Uniprot
Q1XGY6
Q09GR7
Q5MGQ1
I4DKM7
A0A194PXZ7
A0A2H1V6V7
+ More
A0A2A4AQ26 G8FVP5 A0A1E1VZ63 Q25504 A0A2A4J1R7 A0A0K8SWE7 O02443 B9UCR0 A0A290Y5I4 A0A1E1W2P7 A0A0K8TDX9 A0A194R0R0 A0A2A4J2Z2 A0A0L7LAX7 A0A2A4J330 A0A212EXL1 A0A212EXQ7 A0A212EWI0 A0A2H1VIK3 S4S7F7 B3MRZ0 A0A1I8N354 T1PC09 B4JNJ8 C0MLW0 C0MLW5 A0A0L0CAH0 C0H6J7 A0A1I8P830 B4M872 B3NXB8 A0A0Q5THW9 A0A3B0K5Z3 B4HDP4 A0A3B0J9H3 B4F5X8 C0MLW1 B4F5Y6 C0MLV9 Q9VYM0 B4F5X9 Q29J70 B4Q1N4 A0A0Q9XHK0 M9PGZ2 K7IZX0 A0A0R1E9V1 B4L6K0 B4IGG8 C0MLW4 B4NUB9 R4FKX5 T1I377 A0A0M4EJB8 R4G2S2 B4N2G5 A0A1W4V2I2 A0A224XS48 A0A1A9X144 A0A1W4UGQ7 A0A0J7NZ86 B3NSL2 A0A034UYY4 A0A3B0J170 A0A1A9YG66 A0A3F2Z4M3 A0A1B0ABR8 W8AH96 A0A151X6C9 W8BF00 A0A195D5B9 K7IZX1 T1GB68 A0A310SB30 A0A2S1ZS80 Q28YZ2 A0A034VDR9 A0A0K8U746 B4MKZ7 B4MK97 B3MES7 A0A0A1X0X9 B4P7I9 A0A195BCN2
A0A2A4AQ26 G8FVP5 A0A1E1VZ63 Q25504 A0A2A4J1R7 A0A0K8SWE7 O02443 B9UCR0 A0A290Y5I4 A0A1E1W2P7 A0A0K8TDX9 A0A194R0R0 A0A2A4J2Z2 A0A0L7LAX7 A0A2A4J330 A0A212EXL1 A0A212EXQ7 A0A212EWI0 A0A2H1VIK3 S4S7F7 B3MRZ0 A0A1I8N354 T1PC09 B4JNJ8 C0MLW0 C0MLW5 A0A0L0CAH0 C0H6J7 A0A1I8P830 B4M872 B3NXB8 A0A0Q5THW9 A0A3B0K5Z3 B4HDP4 A0A3B0J9H3 B4F5X8 C0MLW1 B4F5Y6 C0MLV9 Q9VYM0 B4F5X9 Q29J70 B4Q1N4 A0A0Q9XHK0 M9PGZ2 K7IZX0 A0A0R1E9V1 B4L6K0 B4IGG8 C0MLW4 B4NUB9 R4FKX5 T1I377 A0A0M4EJB8 R4G2S2 B4N2G5 A0A1W4V2I2 A0A224XS48 A0A1A9X144 A0A1W4UGQ7 A0A0J7NZ86 B3NSL2 A0A034UYY4 A0A3B0J170 A0A1A9YG66 A0A3F2Z4M3 A0A1B0ABR8 W8AH96 A0A151X6C9 W8BF00 A0A195D5B9 K7IZX1 T1GB68 A0A310SB30 A0A2S1ZS80 Q28YZ2 A0A034VDR9 A0A0K8U746 B4MKZ7 B4MK97 B3MES7 A0A0A1X0X9 B4P7I9 A0A195BCN2
Pubmed
EMBL
BABH01013077
AB236661
BR000547
BAE92300.1
FAA00549.1
DQ903306
+ More
ABI55236.1 AY829735 AAV91349.1 AK401845 BAM18467.1 KQ459586 KPI97898.1 ODYU01000983 SOQ36568.1 NWMT01000240 PCC97846.1 JN082715 AER27813.1 GDQN01011036 JAT80018.1 M25486 NWSH01003794 PCG65809.1 GBRD01008194 JAG57627.1 AF004945 EU526837 ACD74813.1 MF318874 ATD87260.1 GDQN01009830 JAT81224.1 GBRD01002102 JAG63719.1 KQ460883 KPJ11383.1 PCG65810.1 JTDY01001912 KOB72554.1 PCG65812.1 AGBW02011718 OWR46238.1 OWR46237.1 AGBW02011965 OWR45860.1 ODYU01002748 SOQ40631.1 JF415081 AFC88814.1 CH902622 EDV34545.2 KA646307 AFP60936.1 CH916371 EDV92291.1 FM246332 CAR94258.1 FM246337 CAR94263.1 JRES01000678 KNC29235.1 BABH01028739 BABH01028740 BR000507 FAA00508.1 CH940653 EDW62348.1 CH954180 EDV47290.1 KQS30439.1 OUUW01000003 SPP78878.1 CH480762 EDW33494.1 SPP78877.1 AM999214 AM999220 AM999223 CAQ53576.1 CAQ53582.1 CAQ53585.1 FM246333 CAR94259.1 AM999222 CAQ53584.1 FM246331 CAR94257.1 AE014298 AY060775 AM999216 AM999217 AM999219 AM999221 FM246327 FM246328 FM246329 FM246330 FM246334 FM246335 AAF48172.1 AAL28323.1 CAQ53578.1 CAQ53579.1 CAQ53581.1 CAQ53583.1 CAR94253.1 CAR94254.1 CAR94255.1 CAR94256.1 CAR94260.1 CAR94261.1 AM999215 AM999218 CAQ53577.1 CAQ53580.1 CH379063 EAL32432.1 CM000162 EDX01475.1 CH933812 KRG07113.1 AGB95328.1 KRK06172.1 EDW05996.1 CH480836 EDW48918.1 FM246336 CAR94262.1 CH983867 EDX16566.1 GAHY01002329 JAA75181.1 ACPB03023185 CP012528 ALC49204.1 GAHY01002351 JAA75159.1 CH963925 EDW78554.1 GFTR01002482 JAW13944.1 LBMM01000707 KMQ97685.1 CH954179 EDV56514.1 GAKP01023459 JAC35499.1 OUUW01000001 SPP74577.1 JXJN01000366 GAMC01018695 JAB87860.1 KQ982482 KYQ55917.1 GAMC01014729 JAB91826.1 KQ976818 KYN08083.1 CAQQ02101294 CAQQ02101295 CAQQ02101296 CAQQ02101297 KQ761824 OAD56752.1 MF942791 AWK28314.1 CM000071 EAL25823.2 GAKP01018723 GAKP01018720 JAC40232.1 GDHF01029911 GDHF01005585 JAI22403.1 JAI46729.1 CH963847 EDW72922.1 CH963846 EDW72536.1 CH902619 EDV35541.1 GBXI01010014 JAD04278.1 CM000158 EDW90024.1 KQ976526 KYM81954.1
ABI55236.1 AY829735 AAV91349.1 AK401845 BAM18467.1 KQ459586 KPI97898.1 ODYU01000983 SOQ36568.1 NWMT01000240 PCC97846.1 JN082715 AER27813.1 GDQN01011036 JAT80018.1 M25486 NWSH01003794 PCG65809.1 GBRD01008194 JAG57627.1 AF004945 EU526837 ACD74813.1 MF318874 ATD87260.1 GDQN01009830 JAT81224.1 GBRD01002102 JAG63719.1 KQ460883 KPJ11383.1 PCG65810.1 JTDY01001912 KOB72554.1 PCG65812.1 AGBW02011718 OWR46238.1 OWR46237.1 AGBW02011965 OWR45860.1 ODYU01002748 SOQ40631.1 JF415081 AFC88814.1 CH902622 EDV34545.2 KA646307 AFP60936.1 CH916371 EDV92291.1 FM246332 CAR94258.1 FM246337 CAR94263.1 JRES01000678 KNC29235.1 BABH01028739 BABH01028740 BR000507 FAA00508.1 CH940653 EDW62348.1 CH954180 EDV47290.1 KQS30439.1 OUUW01000003 SPP78878.1 CH480762 EDW33494.1 SPP78877.1 AM999214 AM999220 AM999223 CAQ53576.1 CAQ53582.1 CAQ53585.1 FM246333 CAR94259.1 AM999222 CAQ53584.1 FM246331 CAR94257.1 AE014298 AY060775 AM999216 AM999217 AM999219 AM999221 FM246327 FM246328 FM246329 FM246330 FM246334 FM246335 AAF48172.1 AAL28323.1 CAQ53578.1 CAQ53579.1 CAQ53581.1 CAQ53583.1 CAR94253.1 CAR94254.1 CAR94255.1 CAR94256.1 CAR94260.1 CAR94261.1 AM999215 AM999218 CAQ53577.1 CAQ53580.1 CH379063 EAL32432.1 CM000162 EDX01475.1 CH933812 KRG07113.1 AGB95328.1 KRK06172.1 EDW05996.1 CH480836 EDW48918.1 FM246336 CAR94262.1 CH983867 EDX16566.1 GAHY01002329 JAA75181.1 ACPB03023185 CP012528 ALC49204.1 GAHY01002351 JAA75159.1 CH963925 EDW78554.1 GFTR01002482 JAW13944.1 LBMM01000707 KMQ97685.1 CH954179 EDV56514.1 GAKP01023459 JAC35499.1 OUUW01000001 SPP74577.1 JXJN01000366 GAMC01018695 JAB87860.1 KQ982482 KYQ55917.1 GAMC01014729 JAB91826.1 KQ976818 KYN08083.1 CAQQ02101294 CAQQ02101295 CAQQ02101296 CAQQ02101297 KQ761824 OAD56752.1 MF942791 AWK28314.1 CM000071 EAL25823.2 GAKP01018723 GAKP01018720 JAC40232.1 GDHF01029911 GDHF01005585 JAI22403.1 JAI46729.1 CH963847 EDW72922.1 CH963846 EDW72536.1 CH902619 EDV35541.1 GBXI01010014 JAD04278.1 CM000158 EDW90024.1 KQ976526 KYM81954.1
Proteomes
UP000005204
UP000053268
UP000243750
UP000218220
UP000053240
UP000037510
+ More
UP000007151 UP000007801 UP000095301 UP000001070 UP000037069 UP000095300 UP000008792 UP000008711 UP000268350 UP000008744 UP000000803 UP000001819 UP000002282 UP000009192 UP000002358 UP000001292 UP000000304 UP000015103 UP000092553 UP000007798 UP000192221 UP000091820 UP000036403 UP000092443 UP000092460 UP000092445 UP000075809 UP000078542 UP000015102 UP000078540
UP000007151 UP000007801 UP000095301 UP000001070 UP000037069 UP000095300 UP000008792 UP000008711 UP000268350 UP000008744 UP000000803 UP000001819 UP000002282 UP000009192 UP000002358 UP000001292 UP000000304 UP000015103 UP000092553 UP000007798 UP000192221 UP000091820 UP000036403 UP000092443 UP000092460 UP000092445 UP000075809 UP000078542 UP000015102 UP000078540
Interpro
SUPFAM
SSF47473
SSF47473
ProteinModelPortal
Q1XGY6
Q09GR7
Q5MGQ1
I4DKM7
A0A194PXZ7
A0A2H1V6V7
+ More
A0A2A4AQ26 G8FVP5 A0A1E1VZ63 Q25504 A0A2A4J1R7 A0A0K8SWE7 O02443 B9UCR0 A0A290Y5I4 A0A1E1W2P7 A0A0K8TDX9 A0A194R0R0 A0A2A4J2Z2 A0A0L7LAX7 A0A2A4J330 A0A212EXL1 A0A212EXQ7 A0A212EWI0 A0A2H1VIK3 S4S7F7 B3MRZ0 A0A1I8N354 T1PC09 B4JNJ8 C0MLW0 C0MLW5 A0A0L0CAH0 C0H6J7 A0A1I8P830 B4M872 B3NXB8 A0A0Q5THW9 A0A3B0K5Z3 B4HDP4 A0A3B0J9H3 B4F5X8 C0MLW1 B4F5Y6 C0MLV9 Q9VYM0 B4F5X9 Q29J70 B4Q1N4 A0A0Q9XHK0 M9PGZ2 K7IZX0 A0A0R1E9V1 B4L6K0 B4IGG8 C0MLW4 B4NUB9 R4FKX5 T1I377 A0A0M4EJB8 R4G2S2 B4N2G5 A0A1W4V2I2 A0A224XS48 A0A1A9X144 A0A1W4UGQ7 A0A0J7NZ86 B3NSL2 A0A034UYY4 A0A3B0J170 A0A1A9YG66 A0A3F2Z4M3 A0A1B0ABR8 W8AH96 A0A151X6C9 W8BF00 A0A195D5B9 K7IZX1 T1GB68 A0A310SB30 A0A2S1ZS80 Q28YZ2 A0A034VDR9 A0A0K8U746 B4MKZ7 B4MK97 B3MES7 A0A0A1X0X9 B4P7I9 A0A195BCN2
A0A2A4AQ26 G8FVP5 A0A1E1VZ63 Q25504 A0A2A4J1R7 A0A0K8SWE7 O02443 B9UCR0 A0A290Y5I4 A0A1E1W2P7 A0A0K8TDX9 A0A194R0R0 A0A2A4J2Z2 A0A0L7LAX7 A0A2A4J330 A0A212EXL1 A0A212EXQ7 A0A212EWI0 A0A2H1VIK3 S4S7F7 B3MRZ0 A0A1I8N354 T1PC09 B4JNJ8 C0MLW0 C0MLW5 A0A0L0CAH0 C0H6J7 A0A1I8P830 B4M872 B3NXB8 A0A0Q5THW9 A0A3B0K5Z3 B4HDP4 A0A3B0J9H3 B4F5X8 C0MLW1 B4F5Y6 C0MLV9 Q9VYM0 B4F5X9 Q29J70 B4Q1N4 A0A0Q9XHK0 M9PGZ2 K7IZX0 A0A0R1E9V1 B4L6K0 B4IGG8 C0MLW4 B4NUB9 R4FKX5 T1I377 A0A0M4EJB8 R4G2S2 B4N2G5 A0A1W4V2I2 A0A224XS48 A0A1A9X144 A0A1W4UGQ7 A0A0J7NZ86 B3NSL2 A0A034UYY4 A0A3B0J170 A0A1A9YG66 A0A3F2Z4M3 A0A1B0ABR8 W8AH96 A0A151X6C9 W8BF00 A0A195D5B9 K7IZX1 T1GB68 A0A310SB30 A0A2S1ZS80 Q28YZ2 A0A034VDR9 A0A0K8U746 B4MKZ7 B4MK97 B3MES7 A0A0A1X0X9 B4P7I9 A0A195BCN2
Ontologies
KEGG
GO
Topology
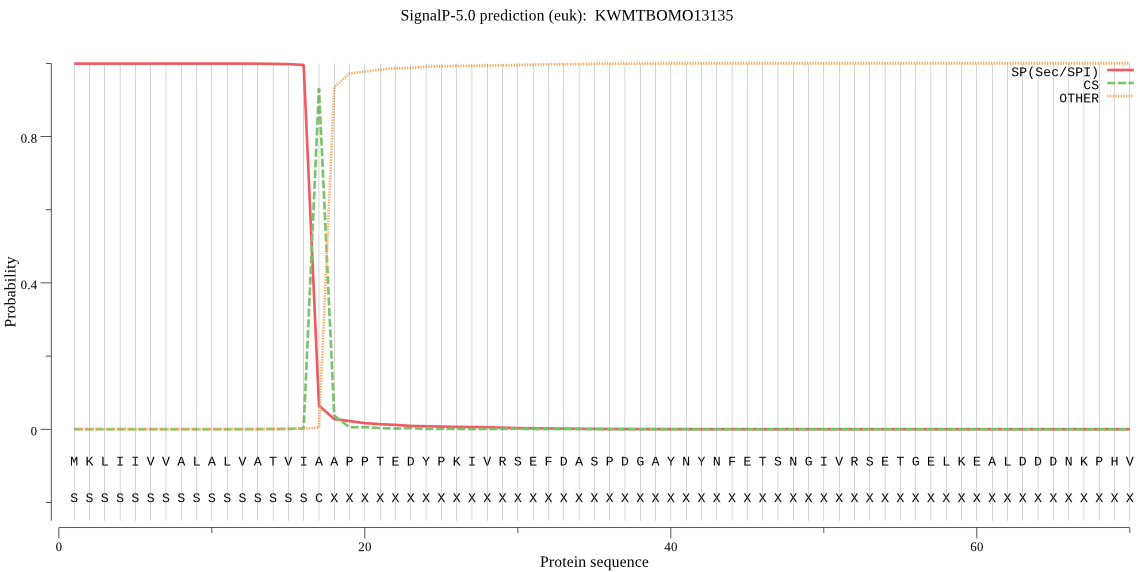
SignalP
Position: 1 - 17,
Likelihood: 0.999078
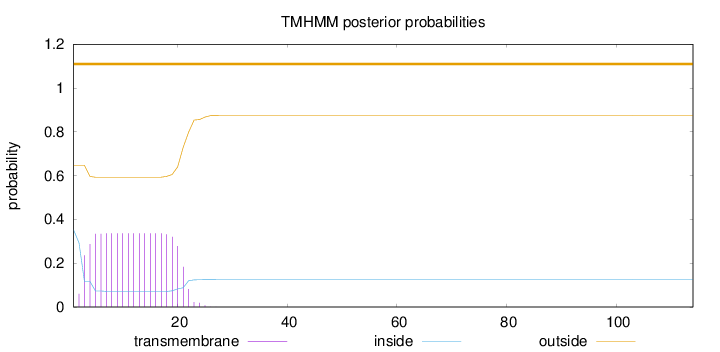
Length:
114
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.19797
Exp number, first 60 AAs:
6.19797
Total prob of N-in:
0.35307
outside
1 - 114
Population Genetic Test Statistics
Pi
226.739567
Theta
200.572114
Tajima's D
0.410603
CLR
0
CSRT
0.48037598120094
Interpretation
Uncertain
