Gene
KWMTBOMO13134
Pre Gene Modal
BGIBMGA000323
Annotation
PREDICTED:_origin_recognition_complex_subunit_3_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.612
Sequence
CDS
ATGGAATCCACTGTGTCAGTTTCAAAGGGAGTGTTTTTATTTCCAAATGGATTTCAAAACTCGAAAACTCTTTCAAAGAAAAAAACAAATTTATTGTTTTGTAAATTATTTCAAGAAGAGTGTTGGTATCTTACTTATAGAAAAAATTGGGAAGCTATTAAAACTAAATTAAATGAACTCAATCAAAGGTCTTACAGTGTGCTGTTGAATGATATAGTAAATTATATAAAGCAGTTTTATGGAAGTGAAGAAGCATTTATGTTGGAAGGAATAATTCCTAGCGCAACACTACTAACTGGGGTTAGTCAGCCTGATCATGTATCACAATTTGATGCACTCATAAATATAATAAAAGATGAAATAAGTCCTCATGTGGCAATAGTTAATTCACAAGATTCGTTATCATTAAAACATATAGTTGAGAACATGGTTTGGCAGTTGATTGGGAATGATTTAAAAAATGACGAAGAATGCGATGTATTATCAACAAAACATAAATTAAAAAAGAGTCAATGTACAATGAAAACCCTGAAAACGTGGTACCATTCAAAATATACAAATGAATCTACTAATAAGAAGAAAAAATCAATTTCGACAAAGACATTAGTAATAGTAATACCTGATTTTGAAAGCTTTAATTGTCACTTGTTACAAGATTTTGTAATGATAATAAGTTCGTACATATCTTCCTTGCCCATAGTGCTTGTTTTTGGGGTGGCAACATCTGTTTCGGCATTGCACAAGTCTTTCCCATATGAGGTGTCATCGAAACTTTTAATAAAAGTTTTTCATTCACATTCGTCTGCGGTTTATATGAATCAGGTGCTAGAAGATATATTTTTAACGCACACAGTTCCATTTCATTTGTCCGGGAAAGCGTTCGAATTGCTCACAGACGTTTTTCTTTTCTACGATTTTTCTGTCAAAGGATTGGTGCAAAGCATTAAGTATTGTATGATGGATCATTTTTATGGTGACAACACAAAGGTCCTCTGTTGTAAGCGTGAAGACCTTGAAGAAGCCATTCTGAATCTAAGTCCGAATGATCTTGAAAGTATCAGACAACTAAAATCGTTCCGACCTTACCTCGAGAAACAAGACTGCCGAACGAAAATAAGTCTTTTTGAAGACGATCAGTATTTCAGGGACGCTCTGTACAAGGAGCTGATGAAATTGCACGATTATTTGTATAGTTTTTATTTGTGCGTGAAATTGTTGTCGACGCTGATCAAGGACATACCGAACAATTTGCTAGGTAAAACGGTACGCGAAGTCTACACGAAATGTGCCACCGAATGCATCACAGACACGCCCTCGTTTAAGGAATGCATGCAGTTACTCAGTTTCCAGTCTCAAATTAAAATCGTCGATACCATCACTGGTGCGTTGAAGTTGCTCAATTCTCATTTACAATCAGTTTCGCCGATCAAACCCCTTCAGGCGACACCGAAGAAGAATAATAATAACACGGATAAAGTGTTTGCACACCGCGAACTGGGCGAAAGCTTCGCGAAGACAGTTCGCGTTCATTTGATGACATTCTTGAGGCAACTCGAAAATGCGAACACTGAAGCTAGTATTAGCACCGCAATGGATACCGACACGAATGACAACGTTGAAAATGTACCGGGAAGTAGATATAAACTGAAAGAGAAATTGTTAAAAGCGACTCGAGTTGAAAAAATTCAATCGGAATTCGAGATGATACGTTGCAGGTTTATAAGTTACCTCGAAGAAATGTTCGCCAAGGGACTCCAGCCTCCGCACACGCAGACGTTTCACGAAATAATGTTCTTTAGTGACATATCGAATGTTAAAAAACACATAGTAGGGGCTCCGCGGGGCGCCCTGCACACCGCGCTCAGCGACCCCGCGCACTATCTGCAGTGCTCGTGCTGTCGGCTTCCGTCGCCGGAGAGCGTGGCCGGAACGCTGCCGGACGTGTGCCTCGCTTACAAGCTGCACCGGGAGTGCGGGAAGCACATCAACCTGTACGACTGGCTGCAAGCCTTCGCAGCCGTCGTCAAGCCCGGGGAAAACGACGAACAACGACATCACGATCCGATCATACAGTCGAGGTTCACCCGCGCGGTGGCCGAATTGCAATTTCTCGGATTTATTAAATCATCAAAGCGAAAGACTGATCACGTAATGCGTTTGACTTGGTAA
Protein
MESTVSVSKGVFLFPNGFQNSKTLSKKKTNLLFCKLFQEECWYLTYRKNWEAIKTKLNELNQRSYSVLLNDIVNYIKQFYGSEEAFMLEGIIPSATLLTGVSQPDHVSQFDALINIIKDEISPHVAIVNSQDSLSLKHIVENMVWQLIGNDLKNDEECDVLSTKHKLKKSQCTMKTLKTWYHSKYTNESTNKKKKSISTKTLVIVIPDFESFNCHLLQDFVMIISSYISSLPIVLVFGVATSVSALHKSFPYEVSSKLLIKVFHSHSSAVYMNQVLEDIFLTHTVPFHLSGKAFELLTDVFLFYDFSVKGLVQSIKYCMMDHFYGDNTKVLCCKREDLEEAILNLSPNDLESIRQLKSFRPYLEKQDCRTKISLFEDDQYFRDALYKELMKLHDYLYSFYLCVKLLSTLIKDIPNNLLGKTVREVYTKCATECITDTPSFKECMQLLSFQSQIKIVDTITGALKLLNSHLQSVSPIKPLQATPKKNNNNTDKVFAHRELGESFAKTVRVHLMTFLRQLENANTEASISTAMDTDTNDNVENVPGSRYKLKEKLLKATRVEKIQSEFEMIRCRFISYLEEMFAKGLQPPHTQTFHEIMFFSDISNVKKHIVGAPRGALHTALSDPAHYLQCSCCRLPSPESVAGTLPDVCLAYKLHRECGKHINLYDWLQAFAAVVKPGENDEQRHHDPIIQSRFTRAVAELQFLGFIKSSKRKTDHVMRLTW
Summary
Uniprot
D3VW58
A0A2A4IXP7
A0A2H1V6Z1
H9ISU6
A0A2A4J297
S4PL05
+ More
A0A194PX47 A0A212EV47 A0A194R0R0 A0A0L7LT89 D6WJ81 A0A2J7QSE1 Q17D50 B4QDZ2 B4HQ18 B4P4T7 Q9Y1B4 B3MDR9 A0A067QSN0 A0A1Q3EW67 Q9TVK9 Q7K2L1 B0XA88 A0A0Q5VMF7 A0A1A9X3B7 A0A182PUJ0 A0A1I8PWI7 A0A0A1X1U0 A0A0M4EWG2 A0A182GV14 A0A1W4UAA9 B4K0G3 B4LIX5 U4U0K6 U4U834 U4TRM0 W5J683 B4GI89 A0A182MTR2 B4N5N7 A0A182QX13 Q28Z58 A0A084VUL6 B4KUE3 A0A034W766 A0A2J7QSF5 A0A0K8U2P8 A0A1A9V3A6 A0A1B0FPQ0 A0A3B0IYH3 A0A1A9XFF2 W8C6E4 A0A1B0AGG4 A0A1B0BAN3 A0A336LPV0 A0A0C9RZS3 A0A1Y1ML64 A0A182IX09 A0A1B6GKL5 A0A0T6BH27 A0A023EVJ2 A0A1Y1ML70 A0A158NB58 A0A154PC10 A0A1Y1MP20 A0A088A0W5 E2BYX6 A0A195EWV6 A0A1Y1ML46 F4WXV9 W5UAE7 A0A1J1HF13 A0A3Q7SDL2 A0A3Q3LDZ4 A0A3Q3QQL3 A0A3Q1II33 A0A384BSW8 A0A3Q7XWI0 A0A3B3DBP2 F1Q348 D2HSL9 K7IY55 A0A3Q1F0B7 A0A091F5L8 A0A3B4WJS8 A0A3B4V0N0 U3JJV8 M3W8W0 A0A091E2V9 A0A3Q7NCM5 J9JYZ8 A0A3Q3FJK4 A0A250Y4F9 U6CV63 A0A3Q1CL30 A0A2H8TNZ5 A0A2Y9L5K4 A0A3Q1F0D2 M3YDF7 Q6TNU7
A0A194PX47 A0A212EV47 A0A194R0R0 A0A0L7LT89 D6WJ81 A0A2J7QSE1 Q17D50 B4QDZ2 B4HQ18 B4P4T7 Q9Y1B4 B3MDR9 A0A067QSN0 A0A1Q3EW67 Q9TVK9 Q7K2L1 B0XA88 A0A0Q5VMF7 A0A1A9X3B7 A0A182PUJ0 A0A1I8PWI7 A0A0A1X1U0 A0A0M4EWG2 A0A182GV14 A0A1W4UAA9 B4K0G3 B4LIX5 U4U0K6 U4U834 U4TRM0 W5J683 B4GI89 A0A182MTR2 B4N5N7 A0A182QX13 Q28Z58 A0A084VUL6 B4KUE3 A0A034W766 A0A2J7QSF5 A0A0K8U2P8 A0A1A9V3A6 A0A1B0FPQ0 A0A3B0IYH3 A0A1A9XFF2 W8C6E4 A0A1B0AGG4 A0A1B0BAN3 A0A336LPV0 A0A0C9RZS3 A0A1Y1ML64 A0A182IX09 A0A1B6GKL5 A0A0T6BH27 A0A023EVJ2 A0A1Y1ML70 A0A158NB58 A0A154PC10 A0A1Y1MP20 A0A088A0W5 E2BYX6 A0A195EWV6 A0A1Y1ML46 F4WXV9 W5UAE7 A0A1J1HF13 A0A3Q7SDL2 A0A3Q3LDZ4 A0A3Q3QQL3 A0A3Q1II33 A0A384BSW8 A0A3Q7XWI0 A0A3B3DBP2 F1Q348 D2HSL9 K7IY55 A0A3Q1F0B7 A0A091F5L8 A0A3B4WJS8 A0A3B4V0N0 U3JJV8 M3W8W0 A0A091E2V9 A0A3Q7NCM5 J9JYZ8 A0A3Q3FJK4 A0A250Y4F9 U6CV63 A0A3Q1CL30 A0A2H8TNZ5 A0A2Y9L5K4 A0A3Q1F0D2 M3YDF7 Q6TNU7
Pubmed
21162711
19121390
23622113
26354079
22118469
26227816
+ More
18362917 19820115 17510324 17994087 22936249 17550304 10346817 24845553 10402192 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25762138 25830018 26483478 23537049 20920257 23761445 15632085 24438588 25348373 24495485 28004739 24945155 21347285 20798317 21719571 23127152 24813606 29451363 16341006 20010809 20075255 17975172 28087693 15520368
18362917 19820115 17510324 17994087 22936249 17550304 10346817 24845553 10402192 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25762138 25830018 26483478 23537049 20920257 23761445 15632085 24438588 25348373 24495485 28004739 24945155 21347285 20798317 21719571 23127152 24813606 29451363 16341006 20010809 20075255 17975172 28087693 15520368
EMBL
GQ214392
ADD10139.1
NWSH01005810
PCG63930.1
ODYU01000983
SOQ36569.1
+ More
BABH01013077 BABH01013078 NWSH01003794 PCG65808.1 GAIX01004275 JAA88285.1 KQ459586 KPI97897.1 AGBW02012251 OWR45356.1 KQ460883 KPJ11383.1 JTDY01000171 KOB78461.1 KQ971343 EFA03157.1 NEVH01011876 PNF31504.1 CH477300 EAT44270.1 CM000362 CM002911 EDX06886.1 KMY93431.1 CH480816 EDW47681.1 CM000158 EDW90658.1 AF139062 AAD39472.1 CH902619 EDV36454.2 KK853069 KDR11812.1 GFDL01015597 JAV19448.1 AF152093 AF153209 AAD32712.2 AAD47349.1 AE013599 AY060817 BT125572 BT132766 BT150363 AAF58411.1 AAL28365.1 AAM68599.1 ADM26240.1 AET07649.1 AGW52173.1 DS232572 EDS43585.1 CH954179 KQS62765.1 GBXI01009567 JAD04725.1 CP012524 ALC42132.1 JXUM01090168 KQ563822 KXJ73230.1 CH916444 EDV95488.1 CH940648 EDW60425.1 KB631120 ERL84121.1 KB631861 ERL86766.1 KB631121 ERL84124.1 ADMH02002024 ETN59932.1 CH479183 EDW36209.1 AXCM01010163 CH964154 EDW79676.1 AXCN02001759 CM000071 EAL25756.1 ATLV01016965 KE525139 KFB41660.1 CH933808 EDW10730.1 GAKP01007536 JAC51416.1 PNF31506.1 GDHF01031689 JAI20625.1 CCAG010016854 OUUW01000001 SPP72885.1 GAMC01001157 JAC05399.1 JXJN01011083 UFQT01000098 SSX19815.1 GBYB01013636 JAG83403.1 GEZM01028150 JAV86429.1 GECZ01006811 JAS62958.1 LJIG01000297 KRT86643.1 GAPW01000425 JAC13173.1 GEZM01028146 JAV86434.1 ADTU01010941 ADTU01010942 KQ434869 KZC09436.1 GEZM01028149 JAV86430.1 GL451531 EFN79094.1 KQ981953 KYN32382.1 GEZM01028147 JAV86433.1 GL888433 EGI60969.1 JT409732 AHH38795.1 CVRI01000001 CRK86551.1 AAEX03008507 GL193291 EFB18841.1 KK719548 KFO64112.1 AGTO01000666 AANG04003966 KN122596 KFO29411.1 ABLF02034108 ABLF02034109 GFFV01001424 JAV38521.1 HAAF01001642 CCP73468.1 GFXV01004071 MBW15876.1 AEYP01067414 AEYP01067415 AEYP01067416 AY391420 AAQ91232.1
BABH01013077 BABH01013078 NWSH01003794 PCG65808.1 GAIX01004275 JAA88285.1 KQ459586 KPI97897.1 AGBW02012251 OWR45356.1 KQ460883 KPJ11383.1 JTDY01000171 KOB78461.1 KQ971343 EFA03157.1 NEVH01011876 PNF31504.1 CH477300 EAT44270.1 CM000362 CM002911 EDX06886.1 KMY93431.1 CH480816 EDW47681.1 CM000158 EDW90658.1 AF139062 AAD39472.1 CH902619 EDV36454.2 KK853069 KDR11812.1 GFDL01015597 JAV19448.1 AF152093 AF153209 AAD32712.2 AAD47349.1 AE013599 AY060817 BT125572 BT132766 BT150363 AAF58411.1 AAL28365.1 AAM68599.1 ADM26240.1 AET07649.1 AGW52173.1 DS232572 EDS43585.1 CH954179 KQS62765.1 GBXI01009567 JAD04725.1 CP012524 ALC42132.1 JXUM01090168 KQ563822 KXJ73230.1 CH916444 EDV95488.1 CH940648 EDW60425.1 KB631120 ERL84121.1 KB631861 ERL86766.1 KB631121 ERL84124.1 ADMH02002024 ETN59932.1 CH479183 EDW36209.1 AXCM01010163 CH964154 EDW79676.1 AXCN02001759 CM000071 EAL25756.1 ATLV01016965 KE525139 KFB41660.1 CH933808 EDW10730.1 GAKP01007536 JAC51416.1 PNF31506.1 GDHF01031689 JAI20625.1 CCAG010016854 OUUW01000001 SPP72885.1 GAMC01001157 JAC05399.1 JXJN01011083 UFQT01000098 SSX19815.1 GBYB01013636 JAG83403.1 GEZM01028150 JAV86429.1 GECZ01006811 JAS62958.1 LJIG01000297 KRT86643.1 GAPW01000425 JAC13173.1 GEZM01028146 JAV86434.1 ADTU01010941 ADTU01010942 KQ434869 KZC09436.1 GEZM01028149 JAV86430.1 GL451531 EFN79094.1 KQ981953 KYN32382.1 GEZM01028147 JAV86433.1 GL888433 EGI60969.1 JT409732 AHH38795.1 CVRI01000001 CRK86551.1 AAEX03008507 GL193291 EFB18841.1 KK719548 KFO64112.1 AGTO01000666 AANG04003966 KN122596 KFO29411.1 ABLF02034108 ABLF02034109 GFFV01001424 JAV38521.1 HAAF01001642 CCP73468.1 GFXV01004071 MBW15876.1 AEYP01067414 AEYP01067415 AEYP01067416 AY391420 AAQ91232.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000007266 UP000235965 UP000008820 UP000000304 UP000001292 UP000002282 UP000007801 UP000027135 UP000000803 UP000002320 UP000008711 UP000091820 UP000075885 UP000095300 UP000092553 UP000069940 UP000249989 UP000192221 UP000001070 UP000008792 UP000030742 UP000000673 UP000008744 UP000075883 UP000007798 UP000075886 UP000001819 UP000030765 UP000009192 UP000078200 UP000092444 UP000268350 UP000092443 UP000092445 UP000092460 UP000075880 UP000005205 UP000076502 UP000005203 UP000008237 UP000078541 UP000007755 UP000221080 UP000183832 UP000286640 UP000261640 UP000261600 UP000265040 UP000261680 UP000291021 UP000286642 UP000261560 UP000002254 UP000002358 UP000257200 UP000052976 UP000261360 UP000261420 UP000016665 UP000011712 UP000028990 UP000286641 UP000007819 UP000261660 UP000257160 UP000248482 UP000000715
UP000007266 UP000235965 UP000008820 UP000000304 UP000001292 UP000002282 UP000007801 UP000027135 UP000000803 UP000002320 UP000008711 UP000091820 UP000075885 UP000095300 UP000092553 UP000069940 UP000249989 UP000192221 UP000001070 UP000008792 UP000030742 UP000000673 UP000008744 UP000075883 UP000007798 UP000075886 UP000001819 UP000030765 UP000009192 UP000078200 UP000092444 UP000268350 UP000092443 UP000092445 UP000092460 UP000075880 UP000005205 UP000076502 UP000005203 UP000008237 UP000078541 UP000007755 UP000221080 UP000183832 UP000286640 UP000261640 UP000261600 UP000265040 UP000261680 UP000291021 UP000286642 UP000261560 UP000002254 UP000002358 UP000257200 UP000052976 UP000261360 UP000261420 UP000016665 UP000011712 UP000028990 UP000286641 UP000007819 UP000261660 UP000257160 UP000248482 UP000000715
Interpro
ProteinModelPortal
D3VW58
A0A2A4IXP7
A0A2H1V6Z1
H9ISU6
A0A2A4J297
S4PL05
+ More
A0A194PX47 A0A212EV47 A0A194R0R0 A0A0L7LT89 D6WJ81 A0A2J7QSE1 Q17D50 B4QDZ2 B4HQ18 B4P4T7 Q9Y1B4 B3MDR9 A0A067QSN0 A0A1Q3EW67 Q9TVK9 Q7K2L1 B0XA88 A0A0Q5VMF7 A0A1A9X3B7 A0A182PUJ0 A0A1I8PWI7 A0A0A1X1U0 A0A0M4EWG2 A0A182GV14 A0A1W4UAA9 B4K0G3 B4LIX5 U4U0K6 U4U834 U4TRM0 W5J683 B4GI89 A0A182MTR2 B4N5N7 A0A182QX13 Q28Z58 A0A084VUL6 B4KUE3 A0A034W766 A0A2J7QSF5 A0A0K8U2P8 A0A1A9V3A6 A0A1B0FPQ0 A0A3B0IYH3 A0A1A9XFF2 W8C6E4 A0A1B0AGG4 A0A1B0BAN3 A0A336LPV0 A0A0C9RZS3 A0A1Y1ML64 A0A182IX09 A0A1B6GKL5 A0A0T6BH27 A0A023EVJ2 A0A1Y1ML70 A0A158NB58 A0A154PC10 A0A1Y1MP20 A0A088A0W5 E2BYX6 A0A195EWV6 A0A1Y1ML46 F4WXV9 W5UAE7 A0A1J1HF13 A0A3Q7SDL2 A0A3Q3LDZ4 A0A3Q3QQL3 A0A3Q1II33 A0A384BSW8 A0A3Q7XWI0 A0A3B3DBP2 F1Q348 D2HSL9 K7IY55 A0A3Q1F0B7 A0A091F5L8 A0A3B4WJS8 A0A3B4V0N0 U3JJV8 M3W8W0 A0A091E2V9 A0A3Q7NCM5 J9JYZ8 A0A3Q3FJK4 A0A250Y4F9 U6CV63 A0A3Q1CL30 A0A2H8TNZ5 A0A2Y9L5K4 A0A3Q1F0D2 M3YDF7 Q6TNU7
A0A194PX47 A0A212EV47 A0A194R0R0 A0A0L7LT89 D6WJ81 A0A2J7QSE1 Q17D50 B4QDZ2 B4HQ18 B4P4T7 Q9Y1B4 B3MDR9 A0A067QSN0 A0A1Q3EW67 Q9TVK9 Q7K2L1 B0XA88 A0A0Q5VMF7 A0A1A9X3B7 A0A182PUJ0 A0A1I8PWI7 A0A0A1X1U0 A0A0M4EWG2 A0A182GV14 A0A1W4UAA9 B4K0G3 B4LIX5 U4U0K6 U4U834 U4TRM0 W5J683 B4GI89 A0A182MTR2 B4N5N7 A0A182QX13 Q28Z58 A0A084VUL6 B4KUE3 A0A034W766 A0A2J7QSF5 A0A0K8U2P8 A0A1A9V3A6 A0A1B0FPQ0 A0A3B0IYH3 A0A1A9XFF2 W8C6E4 A0A1B0AGG4 A0A1B0BAN3 A0A336LPV0 A0A0C9RZS3 A0A1Y1ML64 A0A182IX09 A0A1B6GKL5 A0A0T6BH27 A0A023EVJ2 A0A1Y1ML70 A0A158NB58 A0A154PC10 A0A1Y1MP20 A0A088A0W5 E2BYX6 A0A195EWV6 A0A1Y1ML46 F4WXV9 W5UAE7 A0A1J1HF13 A0A3Q7SDL2 A0A3Q3LDZ4 A0A3Q3QQL3 A0A3Q1II33 A0A384BSW8 A0A3Q7XWI0 A0A3B3DBP2 F1Q348 D2HSL9 K7IY55 A0A3Q1F0B7 A0A091F5L8 A0A3B4WJS8 A0A3B4V0N0 U3JJV8 M3W8W0 A0A091E2V9 A0A3Q7NCM5 J9JYZ8 A0A3Q3FJK4 A0A250Y4F9 U6CV63 A0A3Q1CL30 A0A2H8TNZ5 A0A2Y9L5K4 A0A3Q1F0D2 M3YDF7 Q6TNU7
PDB
4XGC
E-value=1.41748e-135,
Score=1240
Ontologies
GO
PANTHER
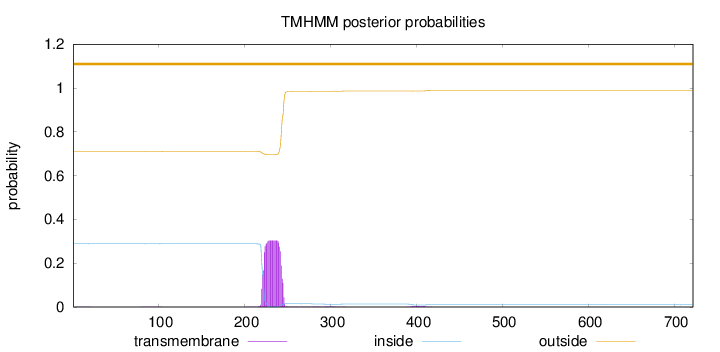
Topology
Length:
722
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.94770999999999
Exp number, first 60 AAs:
0.00505
Total prob of N-in:
0.28851
outside
1 - 722
Population Genetic Test Statistics
Pi
193.545463
Theta
161.270482
Tajima's D
0.508598
CLR
0.204316
CSRT
0.512274386280686
Interpretation
Uncertain
