Gene
KWMTBOMO13131
Annotation
PREDICTED:_uncharacterized_protein_LOC105842558_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.37
Sequence
CDS
ATGCTCAGGGAGGATTCGGTTAGACGTTCAATGCAACCACCTTACTCTGGTCCTCATAGAGTTATATCTAGAGCCGAGGACGGAAAAACGTGGTCGATTGATGTAAAAGGGAAACAGGTGACGGTCAGTGTCGACAGAATTAAGCCAGCTTTCGTCGAGCATTCTTTAGTAGATCTCGTTCCAGCACCAGCACCGCGGGTTTCTGTGCCTCAACCAGCGCAACCGGATGCTACTAATGTCCCTTCATGTCCACTCGTGCCTTCGGCACCTCTAACTGCTACTCCTACACATACACCTCAATATACCACCCGGTCAGGTCGCAGAGTCCACTTCTCCAAACCTTTCGACCTATGA
Protein
MLREDSVRRSMQPPYSGPHRVISRAEDGKTWSIDVKGKQVTVSVDRIKPAFVEHSLVDLVPAPAPRVSVPQPAQPDATNVPSCPLVPSAPLTATPTHTPQYTTRSGRRVHFSKPFDL
Summary
Uniprot
EMBL
GBRD01000337
JAG65484.1
JTDY01005348
KOB67094.1
GBHO01040685
JAG02919.1
+ More
GECZ01031692 JAS38077.1 AAGJ04172791 JTDY01000390 KOB77409.1 IAAA01041607 LAA10577.1 KQ434774 KZC04027.1 NEVH01012562 PNF30270.1 AAGJ04156455 LHQN01021927 KOC58627.1 UZAG01006210 VDO18329.1 NNAY01005137 OXU16960.1 FN356213 CAX83702.1 KK116495 KFM67917.1 EF426679 ABP48077.1 BABH01015766 EU780138 ACI45976.1 KQ434791 KZC05421.1 DF144483 GAA56994.1
GECZ01031692 JAS38077.1 AAGJ04172791 JTDY01000390 KOB77409.1 IAAA01041607 LAA10577.1 KQ434774 KZC04027.1 NEVH01012562 PNF30270.1 AAGJ04156455 LHQN01021927 KOC58627.1 UZAG01006210 VDO18329.1 NNAY01005137 OXU16960.1 FN356213 CAX83702.1 KK116495 KFM67917.1 EF426679 ABP48077.1 BABH01015766 EU780138 ACI45976.1 KQ434791 KZC05421.1 DF144483 GAA56994.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Ontologies
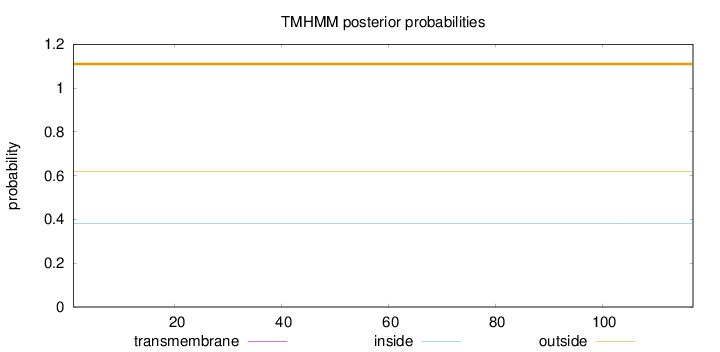
Topology
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0
Total prob of N-in:
0.38140
outside
1 - 117
Population Genetic Test Statistics
Pi
197.100519
Theta
126.771406
Tajima's D
1.639035
CLR
0
CSRT
0.816709164541773
Interpretation
Uncertain
