Gene
KWMTBOMO13129
Annotation
PREDICTED:_uncharacterized_protein_LOC106122315_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 2.555
Sequence
CDS
ATGAAATCGAAATTCGTATACAAAGTACCCTACGATGCAGTGCAGCCCTATACCCGGTTCTCTTCGAAGCTACTAAGCAGGCGCACCAAGATTCGGATATACAAGACTGTAGTCAGACCGATCCTATTGGATGGATGCGAAGCATGGACGCTTACACAGAAAGAAGAGGCAAAACTTCTCGTGACGGAGCGAAAGGTATTGAGAAAAATACTAGGACCAGTTCGGAAAGAAGAAGATTTTTGGCACGTCCGGAAAAATAAGGATGTGGAGAGGCTTTTTGGTGAGCCAAATATTCTGGGGGAGATGAAGGCACGGACGCTCGGATGGCTCGGACATGTAGTACGTATGGAACAGGATCGGGCTGTGAAACGGGTGTACACGGGTGTACCGGAGGGACGCCGCCCCGTAGGGAGGCCCAAGTACCGCCGGTGTGACGCTGTGGAAATAAATTTACGGGAGCTCCAAGTGTCGGACTGGCAAGGTGTTGCGCAGCGCCGGGATGAATGGCAAAGTTTATTGTCGGAGGCCAAGACCCACTTCGGGGTCATAGCGCCAGCTCAGTAG
Protein
MKSKFVYKVPYDAVQPYTRFSSKLLSRRTKIRIYKTVVRPILLDGCEAWTLTQKEEAKLLVTERKVLRKILGPVRKEEDFWHVRKNKDVERLFGEPNILGEMKARTLGWLGHVVRMEQDRAVKRVYTGVPEGRRPVGRPKYRRCDAVEINLRELQVSDWQGVAQRRDEWQSLLSEAKTHFGVIAPAQ
Summary
Uniprot
A0A2P8XYX9
A0A2P8YK05
A0A1B0D0G9
A0A1B6JBJ4
A0A2J7PD61
D7F170
+ More
A0A2P8YFW0 A0A2P8YGJ4 A0A2P8YLJ1 A0A2P8YYC9 A0A1B6HD49 A0A2P8XC08 A0A2P8ZND2 E2ALS0 E2AG07 A0A2P8XUU8 A0A2P8Z9K8 A0A2J7RQE7 A0A2P8Z546 A0A2P8ZCD0 A0A2P8Y7F0 A0A023F0H0 A0A2P8Z9L8 A0A2P8YK77 A0A2P8YW46 A0A2P8XUN2 A0A2P8XKE6 A0A2J7PDA7 A0A023EZ68 A0A2P8ZBA4 A0A2P8XAX7 A0A2P8ZH06 A0A2P8ZIH1 A0A060Q6A2 A0A2P8Z488 A0A2P8YPE1 A0A2P8Z6Z1 A0A2P8XT91 A0A2P8ZFZ0 A0A2P8ZET0 A0A2P8YWA2 A0A2J7QXE7 A0A2P8ZLT6 A0A2P8YGI5 A0A2J7QUH3 A0A2P8YEM8 A0A2P8Y2S1 A0A2P8Z313 A0A034V1D7 A0A2P8YWR2 A0A1B6HHE4 A0A2P8ZBE4 J9M525 A0A2P8ZBE8 A0A0U4HM41 A0A2P8ZEF5 A0A2P8ZAY7 A0A2P8YLD7 A0A2P8ZGH9 A0A2P8YA03 A0A2P8YG08 A0A2P8ZBX0 A0A2P8YU03 A0A2S2PCU1 J9LTW9 A0A2S2N6G4 A0A146KNZ7 A0A367YPS2 J9KWT1 A0A2M4BN64 A0A2J7QTF3 A0A2S2NA50 J9LV30 A0A1B0CZ85 A0A2M4A6U1 A0A2P8Z3H5 A0A1B0EY09 A0A023F0F3 J9KT04 A0A0K8TRS6 A0A2M4A3Y0 A0A2M4A3Z2 A0A0A9YJE1 A0A2P8YIA8 A0A1B0DRK3 A0A2S2PM97 A0A2P8ZMR1 A0A2P8Y7S5 A0A023F0M0 A0A2M4A2X8 A0A2M4A809 A0A2M4ACI4 A0A2P8Z342 A0A2M4A3H0 X1XEZ3 A0A2J7QCQ8 A0A2P8ZDA2 A0A2P8XR62 A0A0A9Y0T5
A0A2P8YFW0 A0A2P8YGJ4 A0A2P8YLJ1 A0A2P8YYC9 A0A1B6HD49 A0A2P8XC08 A0A2P8ZND2 E2ALS0 E2AG07 A0A2P8XUU8 A0A2P8Z9K8 A0A2J7RQE7 A0A2P8Z546 A0A2P8ZCD0 A0A2P8Y7F0 A0A023F0H0 A0A2P8Z9L8 A0A2P8YK77 A0A2P8YW46 A0A2P8XUN2 A0A2P8XKE6 A0A2J7PDA7 A0A023EZ68 A0A2P8ZBA4 A0A2P8XAX7 A0A2P8ZH06 A0A2P8ZIH1 A0A060Q6A2 A0A2P8Z488 A0A2P8YPE1 A0A2P8Z6Z1 A0A2P8XT91 A0A2P8ZFZ0 A0A2P8ZET0 A0A2P8YWA2 A0A2J7QXE7 A0A2P8ZLT6 A0A2P8YGI5 A0A2J7QUH3 A0A2P8YEM8 A0A2P8Y2S1 A0A2P8Z313 A0A034V1D7 A0A2P8YWR2 A0A1B6HHE4 A0A2P8ZBE4 J9M525 A0A2P8ZBE8 A0A0U4HM41 A0A2P8ZEF5 A0A2P8ZAY7 A0A2P8YLD7 A0A2P8ZGH9 A0A2P8YA03 A0A2P8YG08 A0A2P8ZBX0 A0A2P8YU03 A0A2S2PCU1 J9LTW9 A0A2S2N6G4 A0A146KNZ7 A0A367YPS2 J9KWT1 A0A2M4BN64 A0A2J7QTF3 A0A2S2NA50 J9LV30 A0A1B0CZ85 A0A2M4A6U1 A0A2P8Z3H5 A0A1B0EY09 A0A023F0F3 J9KT04 A0A0K8TRS6 A0A2M4A3Y0 A0A2M4A3Z2 A0A0A9YJE1 A0A2P8YIA8 A0A1B0DRK3 A0A2S2PM97 A0A2P8ZMR1 A0A2P8Y7S5 A0A023F0M0 A0A2M4A2X8 A0A2M4A809 A0A2M4ACI4 A0A2P8Z342 A0A2M4A3H0 X1XEZ3 A0A2J7QCQ8 A0A2P8ZDA2 A0A2P8XR62 A0A0A9Y0T5
EMBL
PYGN01001144
PSN37211.1
PYGN01000540
PSN44578.1
AJVK01021402
GECU01011157
+ More
JAS96549.1 NEVH01026393 PNF14276.1 FJ265555 ADI61823.1 PYGN01000629 PSN43121.1 PYGN01000612 PSN43376.1 PYGN01000510 PSN45119.1 PYGN01000287 PSN49259.1 GECU01035086 JAS72620.1 PYGN01003402 PSN29545.1 PYGN01000010 PSN58017.1 GL440612 EFN65619.1 GL439190 EFN67634.1 PYGN01001312 PSN35775.1 PYGN01000136 PSN53183.1 NEVH01001338 PNF43056.1 PYGN01000191 PSN51624.1 PYGN01000106 PSN54154.1 PYGN01000836 PSN40188.1 GBBI01004263 JAC14449.1 PSN53184.1 PYGN01000535 PSN44642.1 PYGN01000325 PSN48468.1 PYGN01001321 PSN35706.1 PYGN01001861 PSN32465.1 NEVH01026392 PNF14311.1 GBBI01004199 JAC14513.1 PYGN01000118 PSN53750.1 PYGN01006068 PSN29155.1 PYGN01000060 PSN55750.1 PYGN01000047 PSN56294.1 HE799691 CCH14900.1 PYGN01000203 PSN51319.1 PYGN01000451 PSN46127.1 PYGN01000168 PSN52268.1 PYGN01001382 PSN35227.1 PYGN01000070 PSN55417.1 PYGN01000078 PSN55011.1 PYGN01000324 PSN48495.1 NEVH01009381 PNF33250.1 PYGN01000020 PSN57453.1 PYGN01000613 PSN43351.1 NEVH01010582 PNF32229.1 PYGN01000657 PSN42717.1 PYGN01001004 PSN38555.1 PYGN01000219 PSN50892.1 GAKP01022638 GAKP01022636 GAKP01022635 JAC36314.1 PYGN01000313 PSN48686.1 GECU01033590 JAS74116.1 PYGN01000116 PSN53825.1 ABLF02024246 PSN53822.1 KU311054 ALX81665.1 PYGN01000080 PSN54879.1 PYGN01000119 PSN53667.1 PYGN01000512 PSN45072.1 PYGN01000064 PSN55609.1 PYGN01000765 PSN41079.1 PYGN01000625 PSN43196.1 PYGN01000111 PSN53993.1 PYGN01000359 PSN47739.1 GGMR01014652 MBY27271.1 ABLF02023718 GGMR01000126 MBY12745.1 GDHC01021627 JAP97001.1 QOUI01000020 RCK67885.1 ABLF02021840 GGFJ01005240 MBW54381.1 NEVH01011195 PNF31853.1 GGMR01001464 MBY14083.1 ABLF02009215 ABLF02009216 AJVK01002125 GGFK01003183 MBW36504.1 PYGN01000212 PSN51056.1 AJVK01015081 GBBI01004261 JAC14451.1 ABLF02035037 GDAI01000742 JAI16861.1 GGFK01002192 MBW35513.1 GGFK01002196 MBW35517.1 GBHO01011285 JAG32319.1 PYGN01000575 PSN43995.1 AJVK01001578 GGMR01017895 MBY30514.1 PYGN01000015 PSN57778.1 PYGN01000827 PSN40309.1 GBBI01004198 JAC14514.1 GGFK01001798 MBW35119.1 GGFK01003613 MBW36934.1 GGFK01005184 MBW38505.1 PYGN01000218 PSN50922.1 GGFK01001949 MBW35270.1 ABLF02032049 ABLF02032052 ABLF02032054 NEVH01016288 PNF26362.1 PYGN01000092 PSN54474.1 PYGN01001492 PSN34499.1 GBHO01016952 JAG26652.1
JAS96549.1 NEVH01026393 PNF14276.1 FJ265555 ADI61823.1 PYGN01000629 PSN43121.1 PYGN01000612 PSN43376.1 PYGN01000510 PSN45119.1 PYGN01000287 PSN49259.1 GECU01035086 JAS72620.1 PYGN01003402 PSN29545.1 PYGN01000010 PSN58017.1 GL440612 EFN65619.1 GL439190 EFN67634.1 PYGN01001312 PSN35775.1 PYGN01000136 PSN53183.1 NEVH01001338 PNF43056.1 PYGN01000191 PSN51624.1 PYGN01000106 PSN54154.1 PYGN01000836 PSN40188.1 GBBI01004263 JAC14449.1 PSN53184.1 PYGN01000535 PSN44642.1 PYGN01000325 PSN48468.1 PYGN01001321 PSN35706.1 PYGN01001861 PSN32465.1 NEVH01026392 PNF14311.1 GBBI01004199 JAC14513.1 PYGN01000118 PSN53750.1 PYGN01006068 PSN29155.1 PYGN01000060 PSN55750.1 PYGN01000047 PSN56294.1 HE799691 CCH14900.1 PYGN01000203 PSN51319.1 PYGN01000451 PSN46127.1 PYGN01000168 PSN52268.1 PYGN01001382 PSN35227.1 PYGN01000070 PSN55417.1 PYGN01000078 PSN55011.1 PYGN01000324 PSN48495.1 NEVH01009381 PNF33250.1 PYGN01000020 PSN57453.1 PYGN01000613 PSN43351.1 NEVH01010582 PNF32229.1 PYGN01000657 PSN42717.1 PYGN01001004 PSN38555.1 PYGN01000219 PSN50892.1 GAKP01022638 GAKP01022636 GAKP01022635 JAC36314.1 PYGN01000313 PSN48686.1 GECU01033590 JAS74116.1 PYGN01000116 PSN53825.1 ABLF02024246 PSN53822.1 KU311054 ALX81665.1 PYGN01000080 PSN54879.1 PYGN01000119 PSN53667.1 PYGN01000512 PSN45072.1 PYGN01000064 PSN55609.1 PYGN01000765 PSN41079.1 PYGN01000625 PSN43196.1 PYGN01000111 PSN53993.1 PYGN01000359 PSN47739.1 GGMR01014652 MBY27271.1 ABLF02023718 GGMR01000126 MBY12745.1 GDHC01021627 JAP97001.1 QOUI01000020 RCK67885.1 ABLF02021840 GGFJ01005240 MBW54381.1 NEVH01011195 PNF31853.1 GGMR01001464 MBY14083.1 ABLF02009215 ABLF02009216 AJVK01002125 GGFK01003183 MBW36504.1 PYGN01000212 PSN51056.1 AJVK01015081 GBBI01004261 JAC14451.1 ABLF02035037 GDAI01000742 JAI16861.1 GGFK01002192 MBW35513.1 GGFK01002196 MBW35517.1 GBHO01011285 JAG32319.1 PYGN01000575 PSN43995.1 AJVK01001578 GGMR01017895 MBY30514.1 PYGN01000015 PSN57778.1 PYGN01000827 PSN40309.1 GBBI01004198 JAC14514.1 GGFK01001798 MBW35119.1 GGFK01003613 MBW36934.1 GGFK01005184 MBW38505.1 PYGN01000218 PSN50922.1 GGFK01001949 MBW35270.1 ABLF02032049 ABLF02032052 ABLF02032054 NEVH01016288 PNF26362.1 PYGN01000092 PSN54474.1 PYGN01001492 PSN34499.1 GBHO01016952 JAG26652.1
Proteomes
PRIDE
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000637 HMGI/Y_DNA-bd_CS
IPR013783 Ig-like_fold
IPR017853 Glycoside_hydrolase_SF
IPR036156 Beta-gal/glucu_dom_sf
IPR006104 Glyco_hydro_2_N
IPR006102 Glyco_hydro_2_Ig-like
IPR041625 Beta-mannosidase_Ig
IPR008979 Galactose-bd-like_sf
IPR041447 Mannosidase_ig
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000637 HMGI/Y_DNA-bd_CS
IPR013783 Ig-like_fold
IPR017853 Glycoside_hydrolase_SF
IPR036156 Beta-gal/glucu_dom_sf
IPR006104 Glyco_hydro_2_N
IPR006102 Glyco_hydro_2_Ig-like
IPR041625 Beta-mannosidase_Ig
IPR008979 Galactose-bd-like_sf
IPR041447 Mannosidase_ig
Gene 3D
ProteinModelPortal
A0A2P8XYX9
A0A2P8YK05
A0A1B0D0G9
A0A1B6JBJ4
A0A2J7PD61
D7F170
+ More
A0A2P8YFW0 A0A2P8YGJ4 A0A2P8YLJ1 A0A2P8YYC9 A0A1B6HD49 A0A2P8XC08 A0A2P8ZND2 E2ALS0 E2AG07 A0A2P8XUU8 A0A2P8Z9K8 A0A2J7RQE7 A0A2P8Z546 A0A2P8ZCD0 A0A2P8Y7F0 A0A023F0H0 A0A2P8Z9L8 A0A2P8YK77 A0A2P8YW46 A0A2P8XUN2 A0A2P8XKE6 A0A2J7PDA7 A0A023EZ68 A0A2P8ZBA4 A0A2P8XAX7 A0A2P8ZH06 A0A2P8ZIH1 A0A060Q6A2 A0A2P8Z488 A0A2P8YPE1 A0A2P8Z6Z1 A0A2P8XT91 A0A2P8ZFZ0 A0A2P8ZET0 A0A2P8YWA2 A0A2J7QXE7 A0A2P8ZLT6 A0A2P8YGI5 A0A2J7QUH3 A0A2P8YEM8 A0A2P8Y2S1 A0A2P8Z313 A0A034V1D7 A0A2P8YWR2 A0A1B6HHE4 A0A2P8ZBE4 J9M525 A0A2P8ZBE8 A0A0U4HM41 A0A2P8ZEF5 A0A2P8ZAY7 A0A2P8YLD7 A0A2P8ZGH9 A0A2P8YA03 A0A2P8YG08 A0A2P8ZBX0 A0A2P8YU03 A0A2S2PCU1 J9LTW9 A0A2S2N6G4 A0A146KNZ7 A0A367YPS2 J9KWT1 A0A2M4BN64 A0A2J7QTF3 A0A2S2NA50 J9LV30 A0A1B0CZ85 A0A2M4A6U1 A0A2P8Z3H5 A0A1B0EY09 A0A023F0F3 J9KT04 A0A0K8TRS6 A0A2M4A3Y0 A0A2M4A3Z2 A0A0A9YJE1 A0A2P8YIA8 A0A1B0DRK3 A0A2S2PM97 A0A2P8ZMR1 A0A2P8Y7S5 A0A023F0M0 A0A2M4A2X8 A0A2M4A809 A0A2M4ACI4 A0A2P8Z342 A0A2M4A3H0 X1XEZ3 A0A2J7QCQ8 A0A2P8ZDA2 A0A2P8XR62 A0A0A9Y0T5
A0A2P8YFW0 A0A2P8YGJ4 A0A2P8YLJ1 A0A2P8YYC9 A0A1B6HD49 A0A2P8XC08 A0A2P8ZND2 E2ALS0 E2AG07 A0A2P8XUU8 A0A2P8Z9K8 A0A2J7RQE7 A0A2P8Z546 A0A2P8ZCD0 A0A2P8Y7F0 A0A023F0H0 A0A2P8Z9L8 A0A2P8YK77 A0A2P8YW46 A0A2P8XUN2 A0A2P8XKE6 A0A2J7PDA7 A0A023EZ68 A0A2P8ZBA4 A0A2P8XAX7 A0A2P8ZH06 A0A2P8ZIH1 A0A060Q6A2 A0A2P8Z488 A0A2P8YPE1 A0A2P8Z6Z1 A0A2P8XT91 A0A2P8ZFZ0 A0A2P8ZET0 A0A2P8YWA2 A0A2J7QXE7 A0A2P8ZLT6 A0A2P8YGI5 A0A2J7QUH3 A0A2P8YEM8 A0A2P8Y2S1 A0A2P8Z313 A0A034V1D7 A0A2P8YWR2 A0A1B6HHE4 A0A2P8ZBE4 J9M525 A0A2P8ZBE8 A0A0U4HM41 A0A2P8ZEF5 A0A2P8ZAY7 A0A2P8YLD7 A0A2P8ZGH9 A0A2P8YA03 A0A2P8YG08 A0A2P8ZBX0 A0A2P8YU03 A0A2S2PCU1 J9LTW9 A0A2S2N6G4 A0A146KNZ7 A0A367YPS2 J9KWT1 A0A2M4BN64 A0A2J7QTF3 A0A2S2NA50 J9LV30 A0A1B0CZ85 A0A2M4A6U1 A0A2P8Z3H5 A0A1B0EY09 A0A023F0F3 J9KT04 A0A0K8TRS6 A0A2M4A3Y0 A0A2M4A3Z2 A0A0A9YJE1 A0A2P8YIA8 A0A1B0DRK3 A0A2S2PM97 A0A2P8ZMR1 A0A2P8Y7S5 A0A023F0M0 A0A2M4A2X8 A0A2M4A809 A0A2M4ACI4 A0A2P8Z342 A0A2M4A3H0 X1XEZ3 A0A2J7QCQ8 A0A2P8ZDA2 A0A2P8XR62 A0A0A9Y0T5
Ontologies
Topology
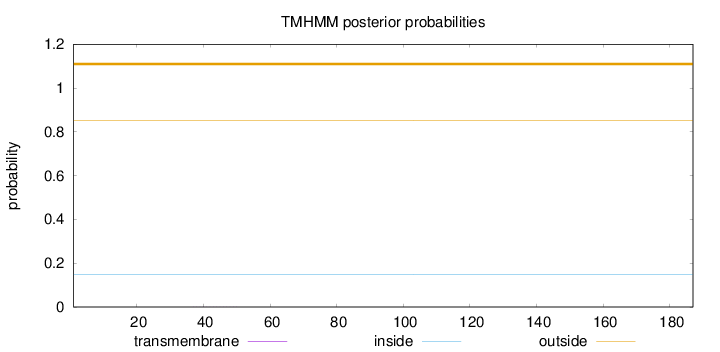
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0025
Exp number, first 60 AAs:
0.00227
Total prob of N-in:
0.14897
outside
1 - 187
Population Genetic Test Statistics
Pi
262.804177
Theta
168.062174
Tajima's D
1.773113
CLR
0
CSRT
0.84390780460977
Interpretation
Uncertain
