Gene
KWMTBOMO13126
Pre Gene Modal
BGIBMGA000375
Annotation
PREDICTED:_tRNA-dihydrouridine(20a/20b)_synthase_[NAD(P)+]-like_[Papilio_machaon]
Full name
tRNA-dihydrouridine synthase
Location in the cell
Cytoplasmic Reliability : 2.303 Extracellular Reliability : 1.825
Sequence
CDS
ATGAAAGAAATCTGCGATATTATGGAAAAATTTGCTGATGCCAAAATAAATAAAACATATTTGAAAGTATGTGCCCCGATGGTCAGATACAGCAAAGTTCATTTTAGAACTTTAGTAAGAAATTTTAATGCAGATCTATGTTTCACTCCAATGATTTTGGCAGATTCTTTTTGTCAAAATTCTAAAGCTAGAGCTAATGAGTTTATGACAACAACAACAGACTTTCCTGTGATAGCCCAATTTGCGGCTAATAATCGAGACGATTTCGTGGATGCTTCAAAACTAGTTTATCCATATGTCGACGGGGTGGACTTAAATTGTGGGTGTCCTCAGAAATGGGCAATGAAAGATGGCTATGGATGTTCCTTATTATCTAAACCAGAAATAATTTATGACATTGTAAAAGGACTAAGAAATGTTTTGCCCAACAAATTTAGTATATCTGTGAAAATAAGACTTTTGAATGATATAAAAAAAACTGTTATTTTGTGCCAGCAGCTAGAGAAATGCGGAGTAACATTTTTGACAATACATGGAAGAACAATTGCACAAAAAAGCAGTGGTCCTGTAGATATAAATGGATTAAAAGAGGTCAGAGAGGCCTTATCTGTCCCAATGGTAGCAAATGGTGGTGTCAAGAATCTTAATGATGCCAACCACTTATATGAAGTAGTAAAGTGTAGTGGTGTAATGGCGGCTGGGGGTCTTTTGTCTAATCCTGCTTTATTCAGTGGTATTACAAAAACTCCACTTAGTTGTGTGAAAATGTGGATGGAATTAAAGAATAAAAACATTGAAAGAATAACATTTCAGTGCTATCATCATCATTTAGTCTTTATGTTAGAAAAAGTATTGACCAAGCAGCAAAAACAAATTTTCAACCATTTAAGTACATTTGAAAGTGTTGATGATTTTATACTGAATAACTTATTTGAGAATTCAATAGTTGATTTTCATTATAAACATGAATTAGGAAACTTTACAGAATGTCAGTTTCCTGATCAAATTACTATGAAACATTGTACTAAATGTAGAACCTGTGGCAAAAGTGTACATTACTGTGTGTGTAATAAATATGACTATAGTACTACCCACGGAAACTTTTTTATCTCGCACATTAATGCAGATGATGGTCTTGATTACATGGACAGTAATATATTTGATGAACCAATATGA
Protein
MKEICDIMEKFADAKINKTYLKVCAPMVRYSKVHFRTLVRNFNADLCFTPMILADSFCQNSKARANEFMTTTTDFPVIAQFAANNRDDFVDASKLVYPYVDGVDLNCGCPQKWAMKDGYGCSLLSKPEIIYDIVKGLRNVLPNKFSISVKIRLLNDIKKTVILCQQLEKCGVTFLTIHGRTIAQKSSGPVDINGLKEVREALSVPMVANGGVKNLNDANHLYEVVKCSGVMAAGGLLSNPALFSGITKTPLSCVKMWMELKNKNIERITFQCYHHHLVFMLEKVLTKQQKQIFNHLSTFESVDDFILNNLFENSIVDFHYKHELGNFTECQFPDQITMKHCTKCRTCGKSVHYCVCNKYDYSTTHGNFFISHINADDGLDYMDSNIFDEPI
Summary
Description
Catalyzes the synthesis of dihydrouridine, a modified base found in the D-loop of most tRNAs.
Cofactor
FMN
Similarity
Belongs to the dus family.
Uniprot
A0A2H1VLL8
A0A3S2TCN7
A0A212EV25
A0A0L7LJX8
A0A182VRV9
A0A182QKF6
+ More
A0A182XVY7 A0A182MTU5 A0A182J8W2 A0A182XIQ9 A0A182KGG2 A0A182HTK6 Q7QCT0 W5JVD0 A0A182LBV0 A0A0K8TLI0 A0A182RGZ9 A0A182H7N5 A0A1B6K4D1 A0A182UH47 Q0IFC6 A0A1B6FT87 A0A182PDY5 A0A182UNV6 A0A182F733 A0A026W4F5 A0A182N799 A0A2M4BR34 A0A1Q3EZA5 A0A1B6M4W7 D6WQ25 A0A0A9YWV7 E9IND1 A0A0C9R4H0 A0A195C7K4 A0A1W4WTJ9 K7IMR7 A0A151WHI5 A0A158NMV5 E2BEE4 A0A232FHI1 A0A023F3T4 A0A195EXU5 A0A154PKR8 A0A2A3EDJ2 A0A0L7QQZ2 E0VKA2 T1HX61 A0A088ASJ4 A0A2M4CLH5 A0A0M9ABK9 J9K6J4 A0A1J1J092 A0A0A9Y6K9 A0A146KV37 A0A2H8TRL2 A0A0K8UY66 A0A2H8TYY1 A0A067R3W9 A0A0A1XBG4 A0A034WG31 A0A1A9WKD7 B0X4B0 A0A1I8M4T1 A0A0B7A037 A0A2J7QMZ2 A0A091NC64 W8C6Q0 A0A087RKP8 E9GJN7 A0A093PC64 A0A093BQG7 A0A091FSM8 K1R069 A0A091HYK6 A0A147BPV2 A0A093PD29 A0A2Y9DX04 A0A091PQY8 A0A131Y2I3 A0A093J7C4 A0A093HBG9 A0A1I8PM34 A0A091K338 A0A3P8WUS4 A0A091IZ79 A0A2I0MH73 A0A091W0D6 A0A093G7E0 A0A0L0CE11 A0A091X9J0 A0A195BC07 G3WHV9 Q29PI6 A0A091NXZ3 A0A0A0ATQ2
A0A182XVY7 A0A182MTU5 A0A182J8W2 A0A182XIQ9 A0A182KGG2 A0A182HTK6 Q7QCT0 W5JVD0 A0A182LBV0 A0A0K8TLI0 A0A182RGZ9 A0A182H7N5 A0A1B6K4D1 A0A182UH47 Q0IFC6 A0A1B6FT87 A0A182PDY5 A0A182UNV6 A0A182F733 A0A026W4F5 A0A182N799 A0A2M4BR34 A0A1Q3EZA5 A0A1B6M4W7 D6WQ25 A0A0A9YWV7 E9IND1 A0A0C9R4H0 A0A195C7K4 A0A1W4WTJ9 K7IMR7 A0A151WHI5 A0A158NMV5 E2BEE4 A0A232FHI1 A0A023F3T4 A0A195EXU5 A0A154PKR8 A0A2A3EDJ2 A0A0L7QQZ2 E0VKA2 T1HX61 A0A088ASJ4 A0A2M4CLH5 A0A0M9ABK9 J9K6J4 A0A1J1J092 A0A0A9Y6K9 A0A146KV37 A0A2H8TRL2 A0A0K8UY66 A0A2H8TYY1 A0A067R3W9 A0A0A1XBG4 A0A034WG31 A0A1A9WKD7 B0X4B0 A0A1I8M4T1 A0A0B7A037 A0A2J7QMZ2 A0A091NC64 W8C6Q0 A0A087RKP8 E9GJN7 A0A093PC64 A0A093BQG7 A0A091FSM8 K1R069 A0A091HYK6 A0A147BPV2 A0A093PD29 A0A2Y9DX04 A0A091PQY8 A0A131Y2I3 A0A093J7C4 A0A093HBG9 A0A1I8PM34 A0A091K338 A0A3P8WUS4 A0A091IZ79 A0A2I0MH73 A0A091W0D6 A0A093G7E0 A0A0L0CE11 A0A091X9J0 A0A195BC07 G3WHV9 Q29PI6 A0A091NXZ3 A0A0A0ATQ2
EC Number
1.3.1.-
Pubmed
22118469
26227816
25244985
12364791
14747013
17210077
+ More
20920257 23761445 20966253 26369729 26483478 17510324 24508170 30249741 18362917 19820115 25401762 21282665 20075255 21347285 20798317 28648823 25474469 20566863 26823975 24845553 25830018 25348373 25315136 24495485 21292972 22992520 29652888 24487278 23371554 26108605 21709235 15632085 17994087
20920257 23761445 20966253 26369729 26483478 17510324 24508170 30249741 18362917 19820115 25401762 21282665 20075255 21347285 20798317 28648823 25474469 20566863 26823975 24845553 25830018 25348373 25315136 24495485 21292972 22992520 29652888 24487278 23371554 26108605 21709235 15632085 17994087
EMBL
ODYU01003232
SOQ41725.1
RSAL01000365
RVE42182.1
AGBW02012251
OWR45350.1
+ More
JTDY01000826 KOB75752.1 AXCN02000147 AXCM01000985 APCN01000539 AAAB01008859 EAA07861.4 ADMH02000237 ETN67283.1 GDAI01002391 JAI15212.1 JXUM01028964 KQ560836 KXJ80677.1 GECU01001442 JAT06265.1 CH477364 EAT42566.1 GECZ01016374 JAS53395.1 KK107432 QOIP01000001 EZA50898.1 RLU27499.1 GGFJ01006404 MBW55545.1 GFDL01014421 JAV20624.1 GEBQ01026423 GEBQ01009010 JAT13554.1 JAT30967.1 KQ971354 EFA06141.2 GBHO01006925 JAG36679.1 GL764295 EFZ17930.1 GBYB01011194 JAG80961.1 KQ978231 KYM96133.1 KQ983115 KYQ47284.1 ADTU01020820 GL447768 EFN85989.1 NNAY01000238 OXU29797.1 GBBI01003133 JAC15579.1 KQ981940 KYN32717.1 KQ434948 KZC12471.1 KZ288271 PBC29778.1 KQ414785 KOC60979.1 AAZO01003005 DS235241 EEB13808.1 ACPB03000369 GGFL01002008 MBW66186.1 KQ435710 KOX79923.1 ABLF02020829 CVRI01000066 CRL05939.1 GBHO01015780 JAG27824.1 GDHC01018471 JAQ00158.1 GFXV01004063 MBW15868.1 GDHF01021054 JAI31260.1 GFXV01007294 MBW19099.1 KK852715 KDR17862.1 GBXI01006489 JAD07803.1 GAKP01005685 JAC53267.1 DS232335 EDS40263.1 HACG01027414 CEK74279.1 NEVH01013202 PNF29947.1 KK828715 KFP74792.1 GAMC01001002 JAC05554.1 KL226413 KFM14052.1 GL732548 EFX80145.1 KL668891 KFW74508.1 KL451477 KFV05990.1 KL447430 KFO73515.1 JH816384 EKC27051.1 KL217905 KFP00265.1 GEGO01003039 JAR92365.1 KL225260 KFW70127.1 KK676997 KFQ09731.1 GEFM01002709 JAP73087.1 KK575025 KFW09838.1 KL205899 KFV76332.1 KK540417 KFP30711.1 KK501259 KFP13667.1 AKCR02000012 PKK29031.1 KL410275 KFQ95293.1 KL214981 KFV62689.1 JRES01000523 KNC30437.1 KK734681 KFR10046.1 KQ976529 KYM81752.1 AEFK01195698 CH379058 EAL34307.1 KK644148 KFP97797.1 KL873146 KGL97939.1
JTDY01000826 KOB75752.1 AXCN02000147 AXCM01000985 APCN01000539 AAAB01008859 EAA07861.4 ADMH02000237 ETN67283.1 GDAI01002391 JAI15212.1 JXUM01028964 KQ560836 KXJ80677.1 GECU01001442 JAT06265.1 CH477364 EAT42566.1 GECZ01016374 JAS53395.1 KK107432 QOIP01000001 EZA50898.1 RLU27499.1 GGFJ01006404 MBW55545.1 GFDL01014421 JAV20624.1 GEBQ01026423 GEBQ01009010 JAT13554.1 JAT30967.1 KQ971354 EFA06141.2 GBHO01006925 JAG36679.1 GL764295 EFZ17930.1 GBYB01011194 JAG80961.1 KQ978231 KYM96133.1 KQ983115 KYQ47284.1 ADTU01020820 GL447768 EFN85989.1 NNAY01000238 OXU29797.1 GBBI01003133 JAC15579.1 KQ981940 KYN32717.1 KQ434948 KZC12471.1 KZ288271 PBC29778.1 KQ414785 KOC60979.1 AAZO01003005 DS235241 EEB13808.1 ACPB03000369 GGFL01002008 MBW66186.1 KQ435710 KOX79923.1 ABLF02020829 CVRI01000066 CRL05939.1 GBHO01015780 JAG27824.1 GDHC01018471 JAQ00158.1 GFXV01004063 MBW15868.1 GDHF01021054 JAI31260.1 GFXV01007294 MBW19099.1 KK852715 KDR17862.1 GBXI01006489 JAD07803.1 GAKP01005685 JAC53267.1 DS232335 EDS40263.1 HACG01027414 CEK74279.1 NEVH01013202 PNF29947.1 KK828715 KFP74792.1 GAMC01001002 JAC05554.1 KL226413 KFM14052.1 GL732548 EFX80145.1 KL668891 KFW74508.1 KL451477 KFV05990.1 KL447430 KFO73515.1 JH816384 EKC27051.1 KL217905 KFP00265.1 GEGO01003039 JAR92365.1 KL225260 KFW70127.1 KK676997 KFQ09731.1 GEFM01002709 JAP73087.1 KK575025 KFW09838.1 KL205899 KFV76332.1 KK540417 KFP30711.1 KK501259 KFP13667.1 AKCR02000012 PKK29031.1 KL410275 KFQ95293.1 KL214981 KFV62689.1 JRES01000523 KNC30437.1 KK734681 KFR10046.1 KQ976529 KYM81752.1 AEFK01195698 CH379058 EAL34307.1 KK644148 KFP97797.1 KL873146 KGL97939.1
Proteomes
UP000283053
UP000007151
UP000037510
UP000075920
UP000075886
UP000076408
+ More
UP000075883 UP000075880 UP000076407 UP000075881 UP000075840 UP000007062 UP000000673 UP000075882 UP000075900 UP000069940 UP000249989 UP000075902 UP000008820 UP000075885 UP000075903 UP000069272 UP000053097 UP000279307 UP000075884 UP000007266 UP000078542 UP000192223 UP000002358 UP000075809 UP000005205 UP000008237 UP000215335 UP000078541 UP000076502 UP000242457 UP000053825 UP000009046 UP000015103 UP000005203 UP000053105 UP000007819 UP000183832 UP000027135 UP000091820 UP000002320 UP000095301 UP000235965 UP000053286 UP000000305 UP000053258 UP000053760 UP000005408 UP000054308 UP000054081 UP000248480 UP000053584 UP000095300 UP000265120 UP000053119 UP000053872 UP000053283 UP000053875 UP000037069 UP000053605 UP000078540 UP000007648 UP000001819 UP000053858
UP000075883 UP000075880 UP000076407 UP000075881 UP000075840 UP000007062 UP000000673 UP000075882 UP000075900 UP000069940 UP000249989 UP000075902 UP000008820 UP000075885 UP000075903 UP000069272 UP000053097 UP000279307 UP000075884 UP000007266 UP000078542 UP000192223 UP000002358 UP000075809 UP000005205 UP000008237 UP000215335 UP000078541 UP000076502 UP000242457 UP000053825 UP000009046 UP000015103 UP000005203 UP000053105 UP000007819 UP000183832 UP000027135 UP000091820 UP000002320 UP000095301 UP000235965 UP000053286 UP000000305 UP000053258 UP000053760 UP000005408 UP000054308 UP000054081 UP000248480 UP000053584 UP000095300 UP000265120 UP000053119 UP000053872 UP000053283 UP000053875 UP000037069 UP000053605 UP000078540 UP000007648 UP000001819 UP000053858
Pfam
PF01207 Dus
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
A0A2H1VLL8
A0A3S2TCN7
A0A212EV25
A0A0L7LJX8
A0A182VRV9
A0A182QKF6
+ More
A0A182XVY7 A0A182MTU5 A0A182J8W2 A0A182XIQ9 A0A182KGG2 A0A182HTK6 Q7QCT0 W5JVD0 A0A182LBV0 A0A0K8TLI0 A0A182RGZ9 A0A182H7N5 A0A1B6K4D1 A0A182UH47 Q0IFC6 A0A1B6FT87 A0A182PDY5 A0A182UNV6 A0A182F733 A0A026W4F5 A0A182N799 A0A2M4BR34 A0A1Q3EZA5 A0A1B6M4W7 D6WQ25 A0A0A9YWV7 E9IND1 A0A0C9R4H0 A0A195C7K4 A0A1W4WTJ9 K7IMR7 A0A151WHI5 A0A158NMV5 E2BEE4 A0A232FHI1 A0A023F3T4 A0A195EXU5 A0A154PKR8 A0A2A3EDJ2 A0A0L7QQZ2 E0VKA2 T1HX61 A0A088ASJ4 A0A2M4CLH5 A0A0M9ABK9 J9K6J4 A0A1J1J092 A0A0A9Y6K9 A0A146KV37 A0A2H8TRL2 A0A0K8UY66 A0A2H8TYY1 A0A067R3W9 A0A0A1XBG4 A0A034WG31 A0A1A9WKD7 B0X4B0 A0A1I8M4T1 A0A0B7A037 A0A2J7QMZ2 A0A091NC64 W8C6Q0 A0A087RKP8 E9GJN7 A0A093PC64 A0A093BQG7 A0A091FSM8 K1R069 A0A091HYK6 A0A147BPV2 A0A093PD29 A0A2Y9DX04 A0A091PQY8 A0A131Y2I3 A0A093J7C4 A0A093HBG9 A0A1I8PM34 A0A091K338 A0A3P8WUS4 A0A091IZ79 A0A2I0MH73 A0A091W0D6 A0A093G7E0 A0A0L0CE11 A0A091X9J0 A0A195BC07 G3WHV9 Q29PI6 A0A091NXZ3 A0A0A0ATQ2
A0A182XVY7 A0A182MTU5 A0A182J8W2 A0A182XIQ9 A0A182KGG2 A0A182HTK6 Q7QCT0 W5JVD0 A0A182LBV0 A0A0K8TLI0 A0A182RGZ9 A0A182H7N5 A0A1B6K4D1 A0A182UH47 Q0IFC6 A0A1B6FT87 A0A182PDY5 A0A182UNV6 A0A182F733 A0A026W4F5 A0A182N799 A0A2M4BR34 A0A1Q3EZA5 A0A1B6M4W7 D6WQ25 A0A0A9YWV7 E9IND1 A0A0C9R4H0 A0A195C7K4 A0A1W4WTJ9 K7IMR7 A0A151WHI5 A0A158NMV5 E2BEE4 A0A232FHI1 A0A023F3T4 A0A195EXU5 A0A154PKR8 A0A2A3EDJ2 A0A0L7QQZ2 E0VKA2 T1HX61 A0A088ASJ4 A0A2M4CLH5 A0A0M9ABK9 J9K6J4 A0A1J1J092 A0A0A9Y6K9 A0A146KV37 A0A2H8TRL2 A0A0K8UY66 A0A2H8TYY1 A0A067R3W9 A0A0A1XBG4 A0A034WG31 A0A1A9WKD7 B0X4B0 A0A1I8M4T1 A0A0B7A037 A0A2J7QMZ2 A0A091NC64 W8C6Q0 A0A087RKP8 E9GJN7 A0A093PC64 A0A093BQG7 A0A091FSM8 K1R069 A0A091HYK6 A0A147BPV2 A0A093PD29 A0A2Y9DX04 A0A091PQY8 A0A131Y2I3 A0A093J7C4 A0A093HBG9 A0A1I8PM34 A0A091K338 A0A3P8WUS4 A0A091IZ79 A0A2I0MH73 A0A091W0D6 A0A093G7E0 A0A0L0CE11 A0A091X9J0 A0A195BC07 G3WHV9 Q29PI6 A0A091NXZ3 A0A0A0ATQ2
PDB
6EZC
E-value=7.14564e-24,
Score=274
Ontologies
GO
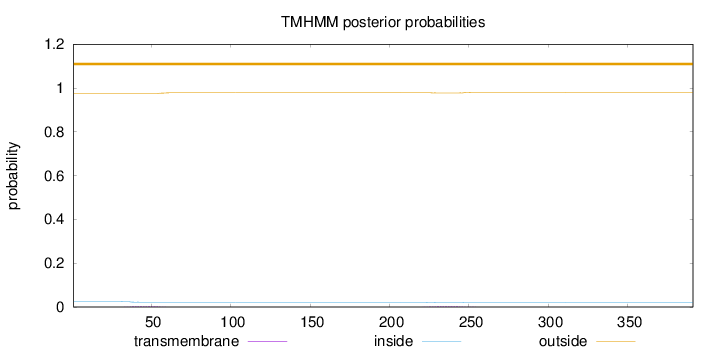
Topology
Length:
391
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1165
Exp number, first 60 AAs:
0.05621
Total prob of N-in:
0.02487
outside
1 - 391
Population Genetic Test Statistics
Pi
238.248688
Theta
203.303407
Tajima's D
0.803017
CLR
0.720346
CSRT
0.608369581520924
Interpretation
Uncertain
