Gene
KWMTBOMO13123
Pre Gene Modal
BGIBMGA000377
Annotation
hypothetical_protein_KGM_16662_[Danaus_plexippus]
Full name
Protein lin-28 homolog
Transcription factor
Location in the cell
Nuclear Reliability : 3.427
Sequence
CDS
ATGAAACTTAATATGTTTAAATTACATTCAGGTGTCGAGGGCGCAAGCGGCAGCGGCAGCGGCGTGACAGGGGTGAAGCGGCGCGGGCGATGCAAGTGGTTCAACGTGGCGAAGGGATGGGGGTTCATTACCCCCGATGACGGCGGCCAAGATGTTTTCGTTCACCAGAGTGTGCTACAAATGCCAGGGTTCAGGTCTCTTGGTGATGAGGAAGTAGAGTTTGAGTGCAAGGAGTCAGATAAGGGCCTGGAAGCGACACGTGTTACAGGTCCCAATGGAACCGATTGTCATGGTTCCGATCGACGACCCTTGTCTAAGATCAGATTCAGGAAGATACGTTGCTACAACTGTGGGGAGTTCGCTAATCACATAGCGGCAAAGTGTAGCATTGGGCCACAACCAAAGAGATGTCACAACTGCAAAAGCGAAGACCACCTCGTTGCTGACTGCCCGATAAAGGTCGAAAAGAAAAAGGATGAACAACAAATGGCTCCGGGCAGCTCCCAAGGAGGAAGCTCACAACAACAAGCGTAA
Protein
MKLNMFKLHSGVEGASGSGSGVTGVKRRGRCKWFNVAKGWGFITPDDGGQDVFVHQSVLQMPGFRSLGDEEVEFECKESDKGLEATRVTGPNGTDCHGSDRRPLSKIRFRKIRCYNCGEFANHIAAKCSIGPQPKRCHNCKSEDHLVADCPIKVEKKKDEQQMAPGSSQGGSSQQQA
Summary
Similarity
Belongs to the lin-28 family.
Keywords
Complete proteome
Cytoplasm
Metal-binding
Reference proteome
Repeat
Zinc
Zinc-finger
Feature
chain Protein lin-28 homolog
Uniprot
H9ISZ8
A0A212EV37
A0A2H1UZW7
A0A0T6BH99
D6WJ86
A0A2P8XTY7
+ More
A0A194R1J8 A0A3S2KZH5 A0A194PXY7 T1JDB5 T1I7Y1 A0A0N7Z9Q0 A0A1B0CV62 T1KM74 K1Q061 A0A1I8MEC8 A0A210QTB8 A0A0L0BTU7 A0A182KGA3 A0A182Y2L5 A0A182FEX1 A0A336K3K3 A0A182MRC5 A0A0Q9XEQ4 A0A0Q9WUJ8 A0A084WMR9 A0A182WBS3 A0A182QNL6 A0A182NIB4 A0A182R8U5 A0A1D2NM73 A0A182JLK4 B0WZK9 A0A0N8ES87 A0A1I8PJ06 Q17DB8 A0A226E215 V3ZTE7 A0A1S3H0S6 A0A1S3H391 A0A1S3H0Y7 A0A034VUL3 A0A0K8V591 A0A0K8W453 R7TPA2 W5JSW1 A0A0P4WYW5 A0A0P5DMH9 A0A0P5EU40 E9GLE5 Q9VRN5 A0A0J9RPV3 A0A1A9W4U5 A0A0P5KSS6 A0A1B0FDU7 A0A1A9Z964 A0A2S2PM89 A0A1A9XSE9 A0A2S2NN77 A0A0P5FUQ7 A0A3B3CG14 A0A3B3DRU1 A0A3B3DSF4 A0A3P9LDG3 A0A3B3IE93 A0A3P9H8U1
A0A194R1J8 A0A3S2KZH5 A0A194PXY7 T1JDB5 T1I7Y1 A0A0N7Z9Q0 A0A1B0CV62 T1KM74 K1Q061 A0A1I8MEC8 A0A210QTB8 A0A0L0BTU7 A0A182KGA3 A0A182Y2L5 A0A182FEX1 A0A336K3K3 A0A182MRC5 A0A0Q9XEQ4 A0A0Q9WUJ8 A0A084WMR9 A0A182WBS3 A0A182QNL6 A0A182NIB4 A0A182R8U5 A0A1D2NM73 A0A182JLK4 B0WZK9 A0A0N8ES87 A0A1I8PJ06 Q17DB8 A0A226E215 V3ZTE7 A0A1S3H0S6 A0A1S3H391 A0A1S3H0Y7 A0A034VUL3 A0A0K8V591 A0A0K8W453 R7TPA2 W5JSW1 A0A0P4WYW5 A0A0P5DMH9 A0A0P5EU40 E9GLE5 Q9VRN5 A0A0J9RPV3 A0A1A9W4U5 A0A0P5KSS6 A0A1B0FDU7 A0A1A9Z964 A0A2S2PM89 A0A1A9XSE9 A0A2S2NN77 A0A0P5FUQ7 A0A3B3CG14 A0A3B3DRU1 A0A3B3DSF4 A0A3P9LDG3 A0A3B3IE93 A0A3P9H8U1
Pubmed
EMBL
BABH01013109
AGBW02012251
OWR45346.1
ODYU01000037
SOQ34131.1
LJIG01000299
+ More
KRT86641.1 KQ971343 EFA04440.2 PYGN01001353 PSN35472.1 KQ460883 KPJ11394.1 RSAL01000365 RVE42179.1 KQ459586 KPI97888.1 JH432102 ACPB03018176 GDRN01136331 JAI56664.1 AJWK01030137 CAEY01000239 JH818227 EKC24739.1 NEDP02002011 OWF51984.1 JRES01001347 KNC23505.1 UFQS01000040 UFQT01000040 SSW98138.1 SSX18524.1 AXCM01010638 CH933809 KRG06137.1 CH940647 KRF84542.1 ATLV01024504 KE525352 KFB51513.1 AXCN02000879 LJIJ01000007 ODN06337.1 DS232209 EDS37523.1 GDUN01000154 JAN95765.1 CH477297 EAT44372.1 LNIX01000009 OXA50516.1 KB203566 ESO84181.1 GAKP01013180 JAC45772.1 GDHF01032750 GDHF01028964 GDHF01018220 GDHF01016360 GDHF01015680 JAI19564.1 JAI23350.1 JAI34094.1 JAI35954.1 JAI36634.1 GDHF01007118 GDHF01006443 JAI45196.1 JAI45871.1 AMQN01002400 KB309137 ELT95392.1 ADMH02000462 ETN66383.1 GDIP01250544 JAI72857.1 GDIP01155206 JAJ68196.1 GDIP01155205 JAJ68197.1 GL732551 EFX79498.1 AF521096 AE014296 AY118360 CM002912 KMY97772.1 GDIQ01185332 JAK66393.1 CCAG010018873 GGMR01017904 MBY30523.1 GGMR01006031 MBY18650.1 GDIQ01251556 JAK00169.1
KRT86641.1 KQ971343 EFA04440.2 PYGN01001353 PSN35472.1 KQ460883 KPJ11394.1 RSAL01000365 RVE42179.1 KQ459586 KPI97888.1 JH432102 ACPB03018176 GDRN01136331 JAI56664.1 AJWK01030137 CAEY01000239 JH818227 EKC24739.1 NEDP02002011 OWF51984.1 JRES01001347 KNC23505.1 UFQS01000040 UFQT01000040 SSW98138.1 SSX18524.1 AXCM01010638 CH933809 KRG06137.1 CH940647 KRF84542.1 ATLV01024504 KE525352 KFB51513.1 AXCN02000879 LJIJ01000007 ODN06337.1 DS232209 EDS37523.1 GDUN01000154 JAN95765.1 CH477297 EAT44372.1 LNIX01000009 OXA50516.1 KB203566 ESO84181.1 GAKP01013180 JAC45772.1 GDHF01032750 GDHF01028964 GDHF01018220 GDHF01016360 GDHF01015680 JAI19564.1 JAI23350.1 JAI34094.1 JAI35954.1 JAI36634.1 GDHF01007118 GDHF01006443 JAI45196.1 JAI45871.1 AMQN01002400 KB309137 ELT95392.1 ADMH02000462 ETN66383.1 GDIP01250544 JAI72857.1 GDIP01155206 JAJ68196.1 GDIP01155205 JAJ68197.1 GL732551 EFX79498.1 AF521096 AE014296 AY118360 CM002912 KMY97772.1 GDIQ01185332 JAK66393.1 CCAG010018873 GGMR01017904 MBY30523.1 GGMR01006031 MBY18650.1 GDIQ01251556 JAK00169.1
Proteomes
UP000005204
UP000007151
UP000007266
UP000245037
UP000053240
UP000283053
+ More
UP000053268 UP000015103 UP000092461 UP000015104 UP000005408 UP000095301 UP000242188 UP000037069 UP000075881 UP000076408 UP000069272 UP000075883 UP000009192 UP000008792 UP000030765 UP000075920 UP000075886 UP000075884 UP000075900 UP000094527 UP000075880 UP000002320 UP000095300 UP000008820 UP000198287 UP000030746 UP000085678 UP000014760 UP000000673 UP000000305 UP000000803 UP000091820 UP000092444 UP000092445 UP000092443 UP000261560 UP000265180 UP000001038 UP000265200
UP000053268 UP000015103 UP000092461 UP000015104 UP000005408 UP000095301 UP000242188 UP000037069 UP000075881 UP000076408 UP000069272 UP000075883 UP000009192 UP000008792 UP000030765 UP000075920 UP000075886 UP000075884 UP000075900 UP000094527 UP000075880 UP000002320 UP000095300 UP000008820 UP000198287 UP000030746 UP000085678 UP000014760 UP000000673 UP000000305 UP000000803 UP000091820 UP000092444 UP000092445 UP000092443 UP000261560 UP000265180 UP000001038 UP000265200
Interpro
CDD
ProteinModelPortal
H9ISZ8
A0A212EV37
A0A2H1UZW7
A0A0T6BH99
D6WJ86
A0A2P8XTY7
+ More
A0A194R1J8 A0A3S2KZH5 A0A194PXY7 T1JDB5 T1I7Y1 A0A0N7Z9Q0 A0A1B0CV62 T1KM74 K1Q061 A0A1I8MEC8 A0A210QTB8 A0A0L0BTU7 A0A182KGA3 A0A182Y2L5 A0A182FEX1 A0A336K3K3 A0A182MRC5 A0A0Q9XEQ4 A0A0Q9WUJ8 A0A084WMR9 A0A182WBS3 A0A182QNL6 A0A182NIB4 A0A182R8U5 A0A1D2NM73 A0A182JLK4 B0WZK9 A0A0N8ES87 A0A1I8PJ06 Q17DB8 A0A226E215 V3ZTE7 A0A1S3H0S6 A0A1S3H391 A0A1S3H0Y7 A0A034VUL3 A0A0K8V591 A0A0K8W453 R7TPA2 W5JSW1 A0A0P4WYW5 A0A0P5DMH9 A0A0P5EU40 E9GLE5 Q9VRN5 A0A0J9RPV3 A0A1A9W4U5 A0A0P5KSS6 A0A1B0FDU7 A0A1A9Z964 A0A2S2PM89 A0A1A9XSE9 A0A2S2NN77 A0A0P5FUQ7 A0A3B3CG14 A0A3B3DRU1 A0A3B3DSF4 A0A3P9LDG3 A0A3B3IE93 A0A3P9H8U1
A0A194R1J8 A0A3S2KZH5 A0A194PXY7 T1JDB5 T1I7Y1 A0A0N7Z9Q0 A0A1B0CV62 T1KM74 K1Q061 A0A1I8MEC8 A0A210QTB8 A0A0L0BTU7 A0A182KGA3 A0A182Y2L5 A0A182FEX1 A0A336K3K3 A0A182MRC5 A0A0Q9XEQ4 A0A0Q9WUJ8 A0A084WMR9 A0A182WBS3 A0A182QNL6 A0A182NIB4 A0A182R8U5 A0A1D2NM73 A0A182JLK4 B0WZK9 A0A0N8ES87 A0A1I8PJ06 Q17DB8 A0A226E215 V3ZTE7 A0A1S3H0S6 A0A1S3H391 A0A1S3H0Y7 A0A034VUL3 A0A0K8V591 A0A0K8W453 R7TPA2 W5JSW1 A0A0P4WYW5 A0A0P5DMH9 A0A0P5EU40 E9GLE5 Q9VRN5 A0A0J9RPV3 A0A1A9W4U5 A0A0P5KSS6 A0A1B0FDU7 A0A1A9Z964 A0A2S2PM89 A0A1A9XSE9 A0A2S2NN77 A0A0P5FUQ7 A0A3B3CG14 A0A3B3DRU1 A0A3B3DSF4 A0A3P9LDG3 A0A3B3IE93 A0A3P9H8U1
PDB
3TS2
E-value=3.44465e-28,
Score=306
Ontologies
GO
Topology
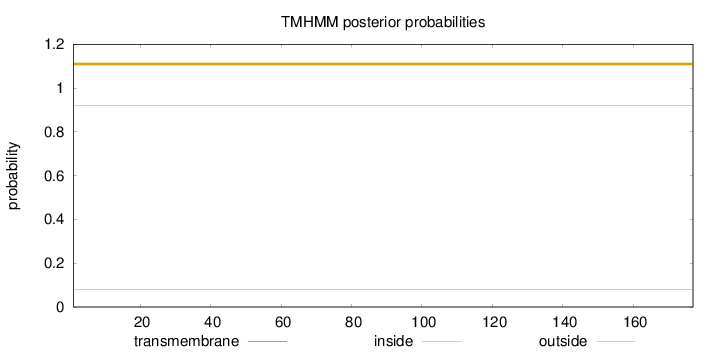
Subcellular location
Cytoplasm
Length:
177
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00112
Exp number, first 60 AAs:
0.00112
Total prob of N-in:
0.07943
outside
1 - 177
Population Genetic Test Statistics
Pi
194.22664
Theta
179.524264
Tajima's D
-0.980412
CLR
0.853754
CSRT
0.143092845357732
Interpretation
Uncertain
