Gene
KWMTBOMO13117 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000381
Annotation
PREDICTED:_nuclear_migration_protein_nudC_isoform_X1_[Bombyx_mori]
Full name
Nuclear migration protein nudC
Alternative Name
Nuclear distribution protein C homolog
Silica-induced gene 92 protein
c15
Silica-induced gene 92 protein
c15
Location in the cell
Nuclear Reliability : 2.823
Sequence
CDS
ATGGCAAACGATCCTGAAAGATTTGATTCCATGCTCCTAGCTATGGCACAACAGCACGAAGGCGGCGTAAAAGATCTGCTGAACACAATAGTTAGCTTTTTGTCTAGAAAAACTGACTTTTTCACTGGTGGCCGTGAAGGGGAATGGGAGCAGGTAGTCAAAGATACTTTCTACACCCATGCTAAAAAAGCATGTGATGAAGCTAATAGGATTAAAAAAGAAAAAGAAGAAGCAGACAAGAGATTAAAAGAGATTCAACAAAGAAAAGAACAAGAGAGACTTGCAAATGATTTTGAACCTGCTACTGTAACTGAACTCACTGATCAGGAAGCAGAGAAGTTAAAGGAAGATATTGAAAAGGACAAAAATGAAAAGAATGCAGAAGTTGGCAGTGGTGATAGCAGTGATGTTACTGAGCCTGCAACGCCCGAAGAAGATGACGATCCCAAAGAAAAGGGGAAGTTGAAACCTAATGCTGGAAATGGATGTGATCTGGAGCACTACAAATGGACACAGACGTTGGAAGAAGTAGAGATTCGAGTGCCGCTGCGACAGATTCTGCGGCCACGCGATCTGACAGTGGTCATCAATAAGAAACACCTCAAGGTTGGCATCAAAGGTCAACCTCTTATAATCGACGGCGAGCTAGATGCCGACGTCAAGATTGAGGAGTCCACGTGGGTGTTGCAGGATGGACGCAATCTACTTATCAACTTAGAAAAGGTGAACAAAATGAACTGGTGGGGTCGGCTTGTTACAACGGACCCAGAAATATCTACAAGGAAAATTAATCCAGAACCATCGAAGCTGTCGGATCTTGATGGTGAAACACGTGGTCTAGTTGAAAAGATGATGTACGATCAGAGACAGAAAGAAATGGGACTACCTACAAGTGATGAACAAAAGAAACAAGAAGTACTCAAGAAATTCATGGAACAGCATCCAGAGATGGACTTCTCCAAATGCAAATTCAACTAA
Protein
MANDPERFDSMLLAMAQQHEGGVKDLLNTIVSFLSRKTDFFTGGREGEWEQVVKDTFYTHAKKACDEANRIKKEKEEADKRLKEIQQRKEQERLANDFEPATVTELTDQEAEKLKEDIEKDKNEKNAEVGSGDSSDVTEPATPEEDDDPKEKGKLKPNAGNGCDLEHYKWTQTLEEVEIRVPLRQILRPRDLTVVINKKHLKVGIKGQPLIIDGELDADVKIEESTWVLQDGRNLLINLEKVNKMNWWGRLVTTDPEISTRKINPEPSKLSDLDGETRGLVEKMMYDQRQKEMGLPTSDEQKKQEVLKKFMEQHPEMDFSKCKFN
Summary
Description
Plays a role in neurogenesis and neuronal migration. Necessary for correct formation of mitotic spindles and chromosome separation during mitosis (By similarity).
Subunit
Interacts with PLK1 (By similarity). Interacts with PAFAH1B1 (PubMed:9601647). Part of a complex containing PLK1, NUDC, dynein and dynactin (PubMed:11734602). Interacts with DCDC1 (By similarity).
Interacts with PAFAH1B1 (PubMed:9601647). Interacts with PLK1 and DCDC1. Part of a complex containing PLK1, NUDC, dynein and dynactin (By similarity).
Interacts with PAFAH1B1 (PubMed:9601647). Interacts with PLK1 and DCDC1. Part of a complex containing PLK1, NUDC, dynein and dynactin (By similarity).
Similarity
Belongs to the nudC family.
Keywords
3D-structure
Acetylation
Cell cycle
Cell division
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Microtubule
Mitosis
Nucleus
Phosphoprotein
Reference proteome
Feature
chain Nuclear migration protein nudC
Uniprot
Q2F5N8
A0A212F9D6
S4P8K9
A0A1E1WIP4
A0A3S2NXJ4
A0A0C9RI57
+ More
A0A1L8DME6 A0A139WHS7 A0A0P5GJ43 A0A162RKR0 K7J0S6 A0A0P5M778 A0A0P5HYJ0 E9H4M9 A0A195EUF6 A0A0P5WGQ6 A0A1B6E9Q9 V5IAR9 A0A1L8DMN4 A0A0P6BC11 L7M6J3 A0A195CKM0 B4LG60 Q29EX0 B4H5M5 A0A0P6CVD6 J3JZL2 A0A1B6L247 A0A067QP89 A0A182J913 A0A2A3EGC0 V9ILC6 J9K0W5 A0A2H8TXV8 Q9VVA6 A0A0M4EXH1 A0A2S2P9V2 A0A1Y1LXM3 B4QMY9 A0A154PCI1 B4KY02 A0A158NK14 B7QH59 U5EVG8 B3NDH2 B4HK70 B4IXG3 A0A131Y236 A0A2S2PVR4 B4N3A7 A0A0K8RPN4 A0A026VV86 A0A3L8DSN6 A0A224XRW0 G3MQQ8 A0A3P8VVJ8 B4PK95 A0A182I1F7 A0A2K5DKP6 A0A1W4WFX2 Q7Q5Z9 A0A182XP68 A0A182KP18 A0A182TJ59 A0A2K6V384 H0WK00 A0A023GIF0 A0A1E1XAL0 A0A0A9X5K3 F7F437 A0A0V0G543 A0A2U3ZA42 G1TEZ4 I3N9X7 A0A096MM99 A0A0D9S835 H9Z4J0 A0A1S3ABV9 A0A2K5RRH3 G1RE19 A0A1S3JUH0 A0A3Q7MWP3 O35685 Q63525 A0A3Q4HX13 A0A250XYI6 B4FBE4 G3RH24 M3YW43 A0A1A6GCZ0 A0A2R8Z5V8 H2PYE7 A0A2K5ZCC5 M3WHP3 A0A2U3VIQ5 A0A061IEW1 A0A3Q4I440 H2N8E2 H9FNY2 A0A2K6GML8
A0A1L8DME6 A0A139WHS7 A0A0P5GJ43 A0A162RKR0 K7J0S6 A0A0P5M778 A0A0P5HYJ0 E9H4M9 A0A195EUF6 A0A0P5WGQ6 A0A1B6E9Q9 V5IAR9 A0A1L8DMN4 A0A0P6BC11 L7M6J3 A0A195CKM0 B4LG60 Q29EX0 B4H5M5 A0A0P6CVD6 J3JZL2 A0A1B6L247 A0A067QP89 A0A182J913 A0A2A3EGC0 V9ILC6 J9K0W5 A0A2H8TXV8 Q9VVA6 A0A0M4EXH1 A0A2S2P9V2 A0A1Y1LXM3 B4QMY9 A0A154PCI1 B4KY02 A0A158NK14 B7QH59 U5EVG8 B3NDH2 B4HK70 B4IXG3 A0A131Y236 A0A2S2PVR4 B4N3A7 A0A0K8RPN4 A0A026VV86 A0A3L8DSN6 A0A224XRW0 G3MQQ8 A0A3P8VVJ8 B4PK95 A0A182I1F7 A0A2K5DKP6 A0A1W4WFX2 Q7Q5Z9 A0A182XP68 A0A182KP18 A0A182TJ59 A0A2K6V384 H0WK00 A0A023GIF0 A0A1E1XAL0 A0A0A9X5K3 F7F437 A0A0V0G543 A0A2U3ZA42 G1TEZ4 I3N9X7 A0A096MM99 A0A0D9S835 H9Z4J0 A0A1S3ABV9 A0A2K5RRH3 G1RE19 A0A1S3JUH0 A0A3Q7MWP3 O35685 Q63525 A0A3Q4HX13 A0A250XYI6 B4FBE4 G3RH24 M3YW43 A0A1A6GCZ0 A0A2R8Z5V8 H2PYE7 A0A2K5ZCC5 M3WHP3 A0A2U3VIQ5 A0A061IEW1 A0A3Q4I440 H2N8E2 H9FNY2 A0A2K6GML8
Pubmed
22118469
23622113
18362917
19820115
20075255
21292972
+ More
25576852 17994087 15632085 22516182 24845553 9376324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 22936249 21347285 24508170 30249741 22216098 24487278 17550304 12364791 14747013 17210077 20966253 28503490 25401762 26823975 25243066 21993624 25319552 7868905 9601647 16141072 15489334 11734602 21183079 7776977 25186727 28087693 19936069 22398555 22722832 16136131 17975172 23929341 29704459
25576852 17994087 15632085 22516182 24845553 9376324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 22936249 21347285 24508170 30249741 22216098 24487278 17550304 12364791 14747013 17210077 20966253 28503490 25401762 26823975 25243066 21993624 25319552 7868905 9601647 16141072 15489334 11734602 21183079 7776977 25186727 28087693 19936069 22398555 22722832 16136131 17975172 23929341 29704459
EMBL
DQ311385
ABD36329.1
AGBW02009593
OWR50365.1
GAIX01004154
JAA88406.1
+ More
GDQN01004323 JAT86731.1 RSAL01000111 RVE47076.1 GBYB01012887 JAG82654.1 GFDF01006448 JAV07636.1 KQ971343 KYB27439.1 GDIQ01239808 JAK11917.1 LRGB01000146 KZS20595.1 GDIQ01159736 JAK91989.1 GDIQ01220913 JAK30812.1 GL732592 EFX73173.1 KQ981979 KYN31532.1 GDIP01086561 JAM17154.1 GEDC01002629 JAS34669.1 GALX01000705 JAB67761.1 GFDF01006413 JAV07671.1 GDIP01017413 JAM86302.1 GACK01005552 JAA59482.1 KQ977642 KYN01002.1 CH940647 EDW69368.1 CH379070 EAL29939.1 CH479211 EDW33077.1 GDIQ01087959 JAN06778.1 BT128288 AEE65523.1 GEBQ01022247 JAT17730.1 KK853097 KDR11435.1 KZ288263 PBC30342.1 JR053182 AEY61870.1 ABLF02024134 ABLF02024135 GFXV01006193 MBW17998.1 AE014296 AY071485 AY328470 AAF49407.2 AAL49107.1 AAP93640.1 AFH04476.1 CP012525 ALC43000.1 GGMR01013349 MBY25968.1 GEZM01045228 JAV77851.1 CM000363 CM002912 EDX10781.1 KMZ00116.1 KQ434870 KZC09605.1 CH933809 EDW18704.1 ADTU01018620 ADTU01018621 ADTU01018622 ADTU01018623 ADTU01018624 ADTU01018625 ADTU01018626 ADTU01018627 ADTU01018628 ADTU01018629 ABJB010242016 ABJB010293257 ABJB010431635 ABJB010582604 ABJB010641009 ABJB010650853 ABJB010826137 ABJB010899687 ABJB010926867 DS937308 EEC18181.1 GANO01001868 JAB58003.1 CH954178 EDV52034.1 CH480815 EDW41811.1 CH916366 EDV96400.1 GEFM01003261 JAP72535.1 GGMS01000231 MBY69434.1 CH964062 EDW78846.1 GADI01000802 JAA73006.1 KK107847 EZA47451.1 QOIP01000005 RLU23183.1 GFTR01005154 JAW11272.1 JO844209 AEO35826.1 CM000159 EDW94793.1 APCN01000055 AAAB01008960 EAA10911.2 AAQR03155822 AAQR03155823 AAQR03155824 AAQR03155825 AAQR03155826 GBBM01001739 JAC33679.1 GFAC01002898 JAT96290.1 GBHO01027607 GBRD01008206 GDHC01000469 JAG15997.1 JAG57615.1 JAQ18160.1 GAMT01006044 GAMR01009208 GAMQ01000006 GAMP01008302 JAB05817.1 JAB24724.1 JAB41845.1 JAB44453.1 GECL01002940 JAP03184.1 AAGW02062843 AAGW02062844 AGTP01007829 AHZZ02015549 AHZZ02015550 AHZZ02015551 AHZZ02015552 AQIB01147820 AQIB01147821 AQIB01147822 JU474052 JV045971 AFH30856.1 AFI36042.1 ADFV01067364 ADFV01067365 ADFV01067366 ADFV01067367 ADFV01067368 X81443 Y15522 AK032673 AK146323 BC011253 X82445 BC065581 GFFV01003488 JAV36457.1 BT034432 ACF79437.1 CABD030001868 AEYP01008124 LZPO01099511 OBS63635.1 AJFE02029023 AJFE02029024 AJFE02029025 AJFE02029026 AJFE02029027 AJFE02029028 AJFE02029029 AACZ04072232 AACZ04072233 AACZ04072234 GABC01000566 GABF01004862 GABD01009136 GABE01009751 NBAG03000438 JAA10772.1 JAA17283.1 JAA23964.1 JAA34988.1 PNI24457.1 AANG04003369 KE668164 RAZU01000087 ERE84178.1 RLQ74456.1 ABGA01131575 ABGA01131576 ABGA01131577 ABGA01131578 ABGA01131579 NDHI03003565 PNJ20494.1 JU332586 AFE76341.1
GDQN01004323 JAT86731.1 RSAL01000111 RVE47076.1 GBYB01012887 JAG82654.1 GFDF01006448 JAV07636.1 KQ971343 KYB27439.1 GDIQ01239808 JAK11917.1 LRGB01000146 KZS20595.1 GDIQ01159736 JAK91989.1 GDIQ01220913 JAK30812.1 GL732592 EFX73173.1 KQ981979 KYN31532.1 GDIP01086561 JAM17154.1 GEDC01002629 JAS34669.1 GALX01000705 JAB67761.1 GFDF01006413 JAV07671.1 GDIP01017413 JAM86302.1 GACK01005552 JAA59482.1 KQ977642 KYN01002.1 CH940647 EDW69368.1 CH379070 EAL29939.1 CH479211 EDW33077.1 GDIQ01087959 JAN06778.1 BT128288 AEE65523.1 GEBQ01022247 JAT17730.1 KK853097 KDR11435.1 KZ288263 PBC30342.1 JR053182 AEY61870.1 ABLF02024134 ABLF02024135 GFXV01006193 MBW17998.1 AE014296 AY071485 AY328470 AAF49407.2 AAL49107.1 AAP93640.1 AFH04476.1 CP012525 ALC43000.1 GGMR01013349 MBY25968.1 GEZM01045228 JAV77851.1 CM000363 CM002912 EDX10781.1 KMZ00116.1 KQ434870 KZC09605.1 CH933809 EDW18704.1 ADTU01018620 ADTU01018621 ADTU01018622 ADTU01018623 ADTU01018624 ADTU01018625 ADTU01018626 ADTU01018627 ADTU01018628 ADTU01018629 ABJB010242016 ABJB010293257 ABJB010431635 ABJB010582604 ABJB010641009 ABJB010650853 ABJB010826137 ABJB010899687 ABJB010926867 DS937308 EEC18181.1 GANO01001868 JAB58003.1 CH954178 EDV52034.1 CH480815 EDW41811.1 CH916366 EDV96400.1 GEFM01003261 JAP72535.1 GGMS01000231 MBY69434.1 CH964062 EDW78846.1 GADI01000802 JAA73006.1 KK107847 EZA47451.1 QOIP01000005 RLU23183.1 GFTR01005154 JAW11272.1 JO844209 AEO35826.1 CM000159 EDW94793.1 APCN01000055 AAAB01008960 EAA10911.2 AAQR03155822 AAQR03155823 AAQR03155824 AAQR03155825 AAQR03155826 GBBM01001739 JAC33679.1 GFAC01002898 JAT96290.1 GBHO01027607 GBRD01008206 GDHC01000469 JAG15997.1 JAG57615.1 JAQ18160.1 GAMT01006044 GAMR01009208 GAMQ01000006 GAMP01008302 JAB05817.1 JAB24724.1 JAB41845.1 JAB44453.1 GECL01002940 JAP03184.1 AAGW02062843 AAGW02062844 AGTP01007829 AHZZ02015549 AHZZ02015550 AHZZ02015551 AHZZ02015552 AQIB01147820 AQIB01147821 AQIB01147822 JU474052 JV045971 AFH30856.1 AFI36042.1 ADFV01067364 ADFV01067365 ADFV01067366 ADFV01067367 ADFV01067368 X81443 Y15522 AK032673 AK146323 BC011253 X82445 BC065581 GFFV01003488 JAV36457.1 BT034432 ACF79437.1 CABD030001868 AEYP01008124 LZPO01099511 OBS63635.1 AJFE02029023 AJFE02029024 AJFE02029025 AJFE02029026 AJFE02029027 AJFE02029028 AJFE02029029 AACZ04072232 AACZ04072233 AACZ04072234 GABC01000566 GABF01004862 GABD01009136 GABE01009751 NBAG03000438 JAA10772.1 JAA17283.1 JAA23964.1 JAA34988.1 PNI24457.1 AANG04003369 KE668164 RAZU01000087 ERE84178.1 RLQ74456.1 ABGA01131575 ABGA01131576 ABGA01131577 ABGA01131578 ABGA01131579 NDHI03003565 PNJ20494.1 JU332586 AFE76341.1
Proteomes
UP000007151
UP000283053
UP000007266
UP000076858
UP000002358
UP000000305
+ More
UP000078541 UP000078542 UP000008792 UP000001819 UP000008744 UP000027135 UP000075880 UP000242457 UP000007819 UP000000803 UP000092553 UP000000304 UP000076502 UP000009192 UP000005205 UP000001555 UP000008711 UP000001292 UP000001070 UP000007798 UP000053097 UP000279307 UP000265120 UP000002282 UP000075840 UP000233020 UP000192223 UP000007062 UP000076407 UP000075882 UP000075902 UP000233220 UP000005225 UP000008225 UP000245340 UP000001811 UP000005215 UP000028761 UP000029965 UP000079721 UP000233040 UP000001073 UP000085678 UP000286641 UP000000589 UP000002494 UP000261580 UP000001519 UP000000715 UP000092124 UP000240080 UP000002277 UP000233140 UP000011712 UP000030759 UP000273346 UP000001595 UP000233160
UP000078541 UP000078542 UP000008792 UP000001819 UP000008744 UP000027135 UP000075880 UP000242457 UP000007819 UP000000803 UP000092553 UP000000304 UP000076502 UP000009192 UP000005205 UP000001555 UP000008711 UP000001292 UP000001070 UP000007798 UP000053097 UP000279307 UP000265120 UP000002282 UP000075840 UP000233020 UP000192223 UP000007062 UP000076407 UP000075882 UP000075902 UP000233220 UP000005225 UP000008225 UP000245340 UP000001811 UP000005215 UP000028761 UP000029965 UP000079721 UP000233040 UP000001073 UP000085678 UP000286641 UP000000589 UP000002494 UP000261580 UP000001519 UP000000715 UP000092124 UP000240080 UP000002277 UP000233140 UP000011712 UP000030759 UP000273346 UP000001595 UP000233160
Interpro
SUPFAM
SSF49764
SSF49764
Gene 3D
ProteinModelPortal
Q2F5N8
A0A212F9D6
S4P8K9
A0A1E1WIP4
A0A3S2NXJ4
A0A0C9RI57
+ More
A0A1L8DME6 A0A139WHS7 A0A0P5GJ43 A0A162RKR0 K7J0S6 A0A0P5M778 A0A0P5HYJ0 E9H4M9 A0A195EUF6 A0A0P5WGQ6 A0A1B6E9Q9 V5IAR9 A0A1L8DMN4 A0A0P6BC11 L7M6J3 A0A195CKM0 B4LG60 Q29EX0 B4H5M5 A0A0P6CVD6 J3JZL2 A0A1B6L247 A0A067QP89 A0A182J913 A0A2A3EGC0 V9ILC6 J9K0W5 A0A2H8TXV8 Q9VVA6 A0A0M4EXH1 A0A2S2P9V2 A0A1Y1LXM3 B4QMY9 A0A154PCI1 B4KY02 A0A158NK14 B7QH59 U5EVG8 B3NDH2 B4HK70 B4IXG3 A0A131Y236 A0A2S2PVR4 B4N3A7 A0A0K8RPN4 A0A026VV86 A0A3L8DSN6 A0A224XRW0 G3MQQ8 A0A3P8VVJ8 B4PK95 A0A182I1F7 A0A2K5DKP6 A0A1W4WFX2 Q7Q5Z9 A0A182XP68 A0A182KP18 A0A182TJ59 A0A2K6V384 H0WK00 A0A023GIF0 A0A1E1XAL0 A0A0A9X5K3 F7F437 A0A0V0G543 A0A2U3ZA42 G1TEZ4 I3N9X7 A0A096MM99 A0A0D9S835 H9Z4J0 A0A1S3ABV9 A0A2K5RRH3 G1RE19 A0A1S3JUH0 A0A3Q7MWP3 O35685 Q63525 A0A3Q4HX13 A0A250XYI6 B4FBE4 G3RH24 M3YW43 A0A1A6GCZ0 A0A2R8Z5V8 H2PYE7 A0A2K5ZCC5 M3WHP3 A0A2U3VIQ5 A0A061IEW1 A0A3Q4I440 H2N8E2 H9FNY2 A0A2K6GML8
A0A1L8DME6 A0A139WHS7 A0A0P5GJ43 A0A162RKR0 K7J0S6 A0A0P5M778 A0A0P5HYJ0 E9H4M9 A0A195EUF6 A0A0P5WGQ6 A0A1B6E9Q9 V5IAR9 A0A1L8DMN4 A0A0P6BC11 L7M6J3 A0A195CKM0 B4LG60 Q29EX0 B4H5M5 A0A0P6CVD6 J3JZL2 A0A1B6L247 A0A067QP89 A0A182J913 A0A2A3EGC0 V9ILC6 J9K0W5 A0A2H8TXV8 Q9VVA6 A0A0M4EXH1 A0A2S2P9V2 A0A1Y1LXM3 B4QMY9 A0A154PCI1 B4KY02 A0A158NK14 B7QH59 U5EVG8 B3NDH2 B4HK70 B4IXG3 A0A131Y236 A0A2S2PVR4 B4N3A7 A0A0K8RPN4 A0A026VV86 A0A3L8DSN6 A0A224XRW0 G3MQQ8 A0A3P8VVJ8 B4PK95 A0A182I1F7 A0A2K5DKP6 A0A1W4WFX2 Q7Q5Z9 A0A182XP68 A0A182KP18 A0A182TJ59 A0A2K6V384 H0WK00 A0A023GIF0 A0A1E1XAL0 A0A0A9X5K3 F7F437 A0A0V0G543 A0A2U3ZA42 G1TEZ4 I3N9X7 A0A096MM99 A0A0D9S835 H9Z4J0 A0A1S3ABV9 A0A2K5RRH3 G1RE19 A0A1S3JUH0 A0A3Q7MWP3 O35685 Q63525 A0A3Q4HX13 A0A250XYI6 B4FBE4 G3RH24 M3YW43 A0A1A6GCZ0 A0A2R8Z5V8 H2PYE7 A0A2K5ZCC5 M3WHP3 A0A2U3VIQ5 A0A061IEW1 A0A3Q4I440 H2N8E2 H9FNY2 A0A2K6GML8
PDB
1WFI
E-value=1.79454e-33,
Score=356
Ontologies
GO
PANTHER
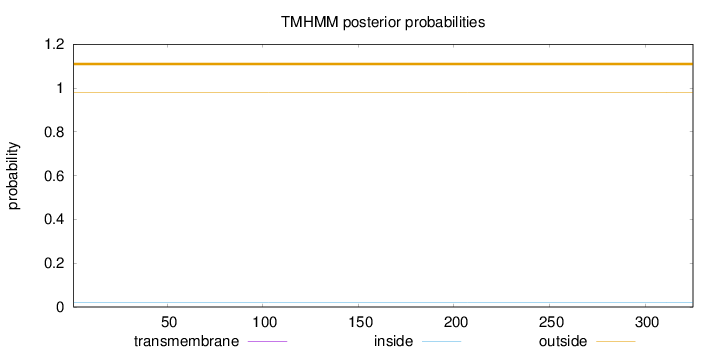
Topology
Subcellular location
Cytoplasm
A small proportion is nuclear, in a punctate pattern (By similarity). In a filamentous pattern adjacent to the nucleus of migrating cerebellar granule cells. Colocalizes with tubulin and dynein and with the microtubule organizing center. Distributed throughout the cytoplasm of non-migrating cells (By similarity). With evidence from 3 publications.
Cytoskeleton A small proportion is nuclear, in a punctate pattern (By similarity). In a filamentous pattern adjacent to the nucleus of migrating cerebellar granule cells. Colocalizes with tubulin and dynein and with the microtubule organizing center. Distributed throughout the cytoplasm of non-migrating cells (By similarity). With evidence from 3 publications.
Nucleus A small proportion is nuclear, in a punctate pattern (By similarity). In a filamentous pattern adjacent to the nucleus of migrating cerebellar granule cells. Colocalizes with tubulin and dynein and with the microtubule organizing center. Distributed throughout the cytoplasm of non-migrating cells (By similarity). With evidence from 3 publications.
Cytoskeleton A small proportion is nuclear, in a punctate pattern (By similarity). In a filamentous pattern adjacent to the nucleus of migrating cerebellar granule cells. Colocalizes with tubulin and dynein and with the microtubule organizing center. Distributed throughout the cytoplasm of non-migrating cells (By similarity). With evidence from 3 publications.
Nucleus A small proportion is nuclear, in a punctate pattern (By similarity). In a filamentous pattern adjacent to the nucleus of migrating cerebellar granule cells. Colocalizes with tubulin and dynein and with the microtubule organizing center. Distributed throughout the cytoplasm of non-migrating cells (By similarity). With evidence from 3 publications.
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00059
Exp number, first 60 AAs:
0.00059
Total prob of N-in:
0.02071
outside
1 - 325
Population Genetic Test Statistics
Pi
226.033396
Theta
191.908687
Tajima's D
0.557334
CLR
0.066139
CSRT
0.531923403829809
Interpretation
Uncertain
