Pre Gene Modal
BGIBMGA000383
Annotation
putative_odorant_response_protein_ODR-4_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.63 PlasmaMembrane Reliability : 1.262
Sequence
CDS
ATGCCCAGAACAGTGTATTCACCCGAGCACTGCCTTCAATATTTGGAACAATATGCCTCCAAAGAAATATTTGCTATAGGGCTAATCCTCGGTCAGATGACCGATGCTAGAGAGAATGTAATCCATCTGGCTCGCACACCAGAAGAAAAAGGATCAGAGATTGGTATAGAATCAAATTATGGCCTAGATAAATCAGAGATAGCAAAGAATTTAAGTAGTGTATCTGAAGCATGGATTGCTGACCATGCGCGACATGTAACTAGAATGCTCCCTGGTGGAATGTTTGTTCAAGGCATATTTGTAACTAGCGATGAAGATGTCTTTGAAGATCCTAATTGCTTTAGTAAATTACGATCAACCCTTAACCACATATATAAAGGCTTAAGTATCAATGAGTATATGTTTGGAAACTGTCCTTACTCTAATGAAAGGCTGATCCTTCACATGTCTACAAGTACTAAAGTACTTACATGCAAAAGCTTAGAAGTCGGAGGTTTGAATAAAAACACAACTTTAAAACCTGTTGAATGGAAATTTTCACCTAAAGCATTGAGTTGGCAAAGGCTGGACTGCTATTATGAGTTTGATGAAATTTATCCTGTTATTGTTAAGAACAGTGGTGTCTCAGTAAAGAATCAATTTCAGCAAATTCTGGAGTCTGCACACAAAATAATTGAATCAAGTGTGATGTTTATTGATGGTGAGCTGAAAGATGGTTCAGAATCATTGGATCAACTCATAAAGAAGAAGAAAACTAAAACAAATACTAAATTGACACAGAATGAAGCACCAAAATCTATGCATGTTTCACTATTTGTACCTTATGAAAACACGCTTCCTGAAACAGTCGAGTATTTAGAATGCGAAGGCAGCATACATTTTAGCGGTGTAGTATCGTCAAGCGTTTTTATATACCCAAAGGCGACAGTAAAAGAGGCGATCGCCGCAGTCAAGCAAGATATTGTTCGTTCACTTGCTTCAAGATTCACAATGCATTGTGATGCATTAATTGATGACAACCTTCTCCCGGAAGAAAAGATTTGTTTCAATGAGCCGCCACGCCGGGTGTTAGTTCCAGTGGGATCCCTTTATCTCTGTGATTACTTATTCCCTGGAGAAGCACCGGCCGAAGCTTTACTTTCTGTTCGGGAACTATTGGATCTAAAAATAACAGAAACTGATGTTATATGCGATGTTGAAACACCTGCAGATACTTCCGAGTTTGACGCCCTAGATCGAGATGCAAGTAGTGAGGAATTGTTGGCAACGCCACAGGAAGCGAGCCAGTTCATGTATATTACGGGACTTTGTTTTGCCATGCTCGTACTCCTTGTTTCTATTATAATTCATTATTACGATGGTATAATTCAAGTGTTGAGTAAAATCTTTCCTAAATCCTGA
Protein
MPRTVYSPEHCLQYLEQYASKEIFAIGLILGQMTDARENVIHLARTPEEKGSEIGIESNYGLDKSEIAKNLSSVSEAWIADHARHVTRMLPGGMFVQGIFVTSDEDVFEDPNCFSKLRSTLNHIYKGLSINEYMFGNCPYSNERLILHMSTSTKVLTCKSLEVGGLNKNTTLKPVEWKFSPKALSWQRLDCYYEFDEIYPVIVKNSGVSVKNQFQQILESAHKIIESSVMFIDGELKDGSESLDQLIKKKKTKTNTKLTQNEAPKSMHVSLFVPYENTLPETVEYLECEGSIHFSGVVSSSVFIYPKATVKEAIAAVKQDIVRSLASRFTMHCDALIDDNLLPEEKICFNEPPRRVLVPVGSLYLCDYLFPGEAPAEALLSVRELLDLKITETDVICDVETPADTSEFDALDRDASSEELLATPQEASQFMYITGLCFAMLVLLVSIIIHYYDGIIQVLSKIFPKS
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A2H1V909
A0A3S2LYI5
A0A2A4J0J9
A0A1E1WLQ0
A0A212F9E9
A0A194PYQ4
+ More
S4PYE3 A0A194R6B6 A0A0L7LCC2 A0A2P8Z8Y6 A0A2J7PFC0 A0A067QRT2 A0A1W4W452 A0A1Y1LZ61 A0A154P1C5 A0A2J7PFC9 E9IPB8 A0A0L7QT15 A0A195DRM3 A0A151I5S5 A0A158NAL2 A0A151WSY4 D6WJD0 F4X280 A0A1W4WFY2 E2A948 A0A2A3EGI2 A0A0C9RHL2 A0A0M9A650 E2BKR4 A0A1B6CVJ3 A0A088AW53 A0A026WAS7 A0A1B6C0Q8 K7IXK9 A0A3L8DGA9 A0A232ET99 A0A1B6KFK8 A0A195EVG9 A0A0J7N3E1 U4TZI2 J3JTQ0 N6U387 A0A0F6YT47 A0A1B6CE79 A0A182KP02 A0A182U1I1 Q7Q5Z7 A0A182I1F5 A0A182V7C4 B0WFM2 A0A182JUT7 Q17KI4 A0A224XG84 A0A1Q3F9Z2 A0A182PDU1 A0A023EWZ3 A0A182XP71 A0A0K8TQR3 E0VB34 A0A0V0G553 A0A1B6HJJ0 A0A069DTR3 A0A023EQ68 A0A0P4VW87 A0A182GGW4 A0A182N8A9 A0A182LYT4 A0A0P4WJT0 A0A182QT88 A0A1S3KCX6 A0A2T7NL92 A0A1S3KCD8 A0A182REY9 A0A1A9WTK3 E9HQF6 A0A0P5RD29 A0A0P5FJS2 A0A131XVY9 A0A162P4D0 B7P873 A0A0P5RY84 A0A0N8ANZ9 A0A2R5LBA6 A0A182STT6 A0A034WG87 L7MEL3 A0A0P4XBC1 A0A182Y2T4 A0A182J7F1 A0A224YS15 A0A2M4BNJ3 A0A0P5N6L6 A0A1I8M4H6 T1PEN9 A0A0L0C3N8
S4PYE3 A0A194R6B6 A0A0L7LCC2 A0A2P8Z8Y6 A0A2J7PFC0 A0A067QRT2 A0A1W4W452 A0A1Y1LZ61 A0A154P1C5 A0A2J7PFC9 E9IPB8 A0A0L7QT15 A0A195DRM3 A0A151I5S5 A0A158NAL2 A0A151WSY4 D6WJD0 F4X280 A0A1W4WFY2 E2A948 A0A2A3EGI2 A0A0C9RHL2 A0A0M9A650 E2BKR4 A0A1B6CVJ3 A0A088AW53 A0A026WAS7 A0A1B6C0Q8 K7IXK9 A0A3L8DGA9 A0A232ET99 A0A1B6KFK8 A0A195EVG9 A0A0J7N3E1 U4TZI2 J3JTQ0 N6U387 A0A0F6YT47 A0A1B6CE79 A0A182KP02 A0A182U1I1 Q7Q5Z7 A0A182I1F5 A0A182V7C4 B0WFM2 A0A182JUT7 Q17KI4 A0A224XG84 A0A1Q3F9Z2 A0A182PDU1 A0A023EWZ3 A0A182XP71 A0A0K8TQR3 E0VB34 A0A0V0G553 A0A1B6HJJ0 A0A069DTR3 A0A023EQ68 A0A0P4VW87 A0A182GGW4 A0A182N8A9 A0A182LYT4 A0A0P4WJT0 A0A182QT88 A0A1S3KCX6 A0A2T7NL92 A0A1S3KCD8 A0A182REY9 A0A1A9WTK3 E9HQF6 A0A0P5RD29 A0A0P5FJS2 A0A131XVY9 A0A162P4D0 B7P873 A0A0P5RY84 A0A0N8ANZ9 A0A2R5LBA6 A0A182STT6 A0A034WG87 L7MEL3 A0A0P4XBC1 A0A182Y2T4 A0A182J7F1 A0A224YS15 A0A2M4BNJ3 A0A0P5N6L6 A0A1I8M4H6 T1PEN9 A0A0L0C3N8
Pubmed
22118469
26354079
23622113
26227816
29403074
24845553
+ More
28004739 21282665 21347285 18362917 19820115 21719571 20798317 24508170 20075255 30249741 28648823 23537049 22516182 20966253 12364791 14747013 17210077 17510324 25474469 26369729 20566863 26334808 24945155 27129103 26483478 21292972 25348373 25576852 25244985 28797301 25315136 26108605
28004739 21282665 21347285 18362917 19820115 21719571 20798317 24508170 20075255 30249741 28648823 23537049 22516182 20966253 12364791 14747013 17210077 17510324 25474469 26369729 20566863 26334808 24945155 27129103 26483478 21292972 25348373 25576852 25244985 28797301 25315136 26108605
EMBL
ODYU01001290
SOQ37299.1
RSAL01000111
RVE47078.1
NWSH01004177
PCG65399.1
+ More
GDQN01003176 JAT87878.1 AGBW02009593 OWR50364.1 KQ459586 KPI97879.1 GAIX01003438 JAA89122.1 KQ460883 KPJ11406.1 JTDY01001802 KOB72846.1 PYGN01000143 PSN52959.1 NEVH01025672 PNF15033.1 KK853097 KDR11437.1 GEZM01043736 GEZM01043735 GEZM01043734 JAV78872.1 KQ434786 KZC05038.1 PNF15035.1 GL764479 EFZ17578.1 KQ414755 KOC61793.1 KQ980612 KYN15149.1 KQ976413 KYM90373.1 ADTU01010371 KQ982779 KYQ50735.1 KQ971343 EFA04456.1 GL888565 EGI59496.1 GL437711 EFN70033.1 KZ288263 PBC30329.1 GBYB01006421 JAG76188.1 KQ435727 KOX78020.1 GL448819 EFN83762.1 GEDC01019788 JAS17510.1 KK107320 EZA52781.1 GEDC01030197 JAS07101.1 AAZX01000028 QOIP01000009 RLU18949.1 NNAY01002333 OXU21526.1 GEBQ01029729 JAT10248.1 KQ981954 KYN32255.1 LBMM01010792 KMQ87230.1 KB631695 ERL85423.1 BT126607 AEE61571.1 APGK01051520 KB741190 ENN73037.1 KM527951 AKF17721.1 GEDC01025556 JAS11742.1 AAAB01008960 EAA11269.4 APCN01000054 DS231919 EDS26328.1 CH477224 EAT47167.1 GFTR01006382 JAW10044.1 GFDL01010651 JAV24394.1 GBBI01005004 JAC13708.1 GDAI01001102 JAI16501.1 DS235021 EEB10590.1 GECL01002955 JAP03169.1 GECU01032951 JAS74755.1 GBGD01001421 JAC87468.1 GAPW01001886 JAC11712.1 GDKW01000226 JAI56369.1 JXUM01062578 KQ562206 KXJ76417.1 AXCM01001908 GDRN01036627 JAI67334.1 AXCN02000964 PZQS01000011 PVD21934.1 GL732719 EFX66031.1 GDIQ01102502 JAL49224.1 GDIQ01253559 JAJ98165.1 GEFM01005960 JAP69836.1 LRGB01000512 KZS18509.1 ABJB010584201 ABJB011026942 DS656359 EEC02795.1 GDIQ01094742 JAL56984.1 GDIQ01257067 JAJ94657.1 GGLE01002685 MBY06811.1 GAKP01006179 JAC52773.1 GACK01003425 JAA61609.1 GDIP01244306 JAI79095.1 GFPF01005454 MAA16600.1 GGFJ01005392 MBW54533.1 GDIQ01157878 JAK93847.1 KA647164 AFP61793.1 JRES01000951 KNC26862.1
GDQN01003176 JAT87878.1 AGBW02009593 OWR50364.1 KQ459586 KPI97879.1 GAIX01003438 JAA89122.1 KQ460883 KPJ11406.1 JTDY01001802 KOB72846.1 PYGN01000143 PSN52959.1 NEVH01025672 PNF15033.1 KK853097 KDR11437.1 GEZM01043736 GEZM01043735 GEZM01043734 JAV78872.1 KQ434786 KZC05038.1 PNF15035.1 GL764479 EFZ17578.1 KQ414755 KOC61793.1 KQ980612 KYN15149.1 KQ976413 KYM90373.1 ADTU01010371 KQ982779 KYQ50735.1 KQ971343 EFA04456.1 GL888565 EGI59496.1 GL437711 EFN70033.1 KZ288263 PBC30329.1 GBYB01006421 JAG76188.1 KQ435727 KOX78020.1 GL448819 EFN83762.1 GEDC01019788 JAS17510.1 KK107320 EZA52781.1 GEDC01030197 JAS07101.1 AAZX01000028 QOIP01000009 RLU18949.1 NNAY01002333 OXU21526.1 GEBQ01029729 JAT10248.1 KQ981954 KYN32255.1 LBMM01010792 KMQ87230.1 KB631695 ERL85423.1 BT126607 AEE61571.1 APGK01051520 KB741190 ENN73037.1 KM527951 AKF17721.1 GEDC01025556 JAS11742.1 AAAB01008960 EAA11269.4 APCN01000054 DS231919 EDS26328.1 CH477224 EAT47167.1 GFTR01006382 JAW10044.1 GFDL01010651 JAV24394.1 GBBI01005004 JAC13708.1 GDAI01001102 JAI16501.1 DS235021 EEB10590.1 GECL01002955 JAP03169.1 GECU01032951 JAS74755.1 GBGD01001421 JAC87468.1 GAPW01001886 JAC11712.1 GDKW01000226 JAI56369.1 JXUM01062578 KQ562206 KXJ76417.1 AXCM01001908 GDRN01036627 JAI67334.1 AXCN02000964 PZQS01000011 PVD21934.1 GL732719 EFX66031.1 GDIQ01102502 JAL49224.1 GDIQ01253559 JAJ98165.1 GEFM01005960 JAP69836.1 LRGB01000512 KZS18509.1 ABJB010584201 ABJB011026942 DS656359 EEC02795.1 GDIQ01094742 JAL56984.1 GDIQ01257067 JAJ94657.1 GGLE01002685 MBY06811.1 GAKP01006179 JAC52773.1 GACK01003425 JAA61609.1 GDIP01244306 JAI79095.1 GFPF01005454 MAA16600.1 GGFJ01005392 MBW54533.1 GDIQ01157878 JAK93847.1 KA647164 AFP61793.1 JRES01000951 KNC26862.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000245037 UP000235965 UP000027135 UP000192223 UP000076502 UP000053825 UP000078492 UP000078540 UP000005205 UP000075809 UP000007266 UP000007755 UP000000311 UP000242457 UP000053105 UP000008237 UP000005203 UP000053097 UP000002358 UP000279307 UP000215335 UP000078541 UP000036403 UP000030742 UP000019118 UP000075882 UP000075902 UP000007062 UP000075840 UP000075903 UP000002320 UP000075881 UP000008820 UP000075885 UP000076407 UP000009046 UP000069940 UP000249989 UP000075884 UP000075883 UP000075886 UP000085678 UP000245119 UP000075900 UP000091820 UP000000305 UP000076858 UP000001555 UP000075901 UP000076408 UP000075880 UP000095301 UP000037069
UP000245037 UP000235965 UP000027135 UP000192223 UP000076502 UP000053825 UP000078492 UP000078540 UP000005205 UP000075809 UP000007266 UP000007755 UP000000311 UP000242457 UP000053105 UP000008237 UP000005203 UP000053097 UP000002358 UP000279307 UP000215335 UP000078541 UP000036403 UP000030742 UP000019118 UP000075882 UP000075902 UP000007062 UP000075840 UP000075903 UP000002320 UP000075881 UP000008820 UP000075885 UP000076407 UP000009046 UP000069940 UP000249989 UP000075884 UP000075883 UP000075886 UP000085678 UP000245119 UP000075900 UP000091820 UP000000305 UP000076858 UP000001555 UP000075901 UP000076408 UP000075880 UP000095301 UP000037069
Interpro
SUPFAM
SSF50494
SSF50494
Gene 3D
CDD
ProteinModelPortal
A0A2H1V909
A0A3S2LYI5
A0A2A4J0J9
A0A1E1WLQ0
A0A212F9E9
A0A194PYQ4
+ More
S4PYE3 A0A194R6B6 A0A0L7LCC2 A0A2P8Z8Y6 A0A2J7PFC0 A0A067QRT2 A0A1W4W452 A0A1Y1LZ61 A0A154P1C5 A0A2J7PFC9 E9IPB8 A0A0L7QT15 A0A195DRM3 A0A151I5S5 A0A158NAL2 A0A151WSY4 D6WJD0 F4X280 A0A1W4WFY2 E2A948 A0A2A3EGI2 A0A0C9RHL2 A0A0M9A650 E2BKR4 A0A1B6CVJ3 A0A088AW53 A0A026WAS7 A0A1B6C0Q8 K7IXK9 A0A3L8DGA9 A0A232ET99 A0A1B6KFK8 A0A195EVG9 A0A0J7N3E1 U4TZI2 J3JTQ0 N6U387 A0A0F6YT47 A0A1B6CE79 A0A182KP02 A0A182U1I1 Q7Q5Z7 A0A182I1F5 A0A182V7C4 B0WFM2 A0A182JUT7 Q17KI4 A0A224XG84 A0A1Q3F9Z2 A0A182PDU1 A0A023EWZ3 A0A182XP71 A0A0K8TQR3 E0VB34 A0A0V0G553 A0A1B6HJJ0 A0A069DTR3 A0A023EQ68 A0A0P4VW87 A0A182GGW4 A0A182N8A9 A0A182LYT4 A0A0P4WJT0 A0A182QT88 A0A1S3KCX6 A0A2T7NL92 A0A1S3KCD8 A0A182REY9 A0A1A9WTK3 E9HQF6 A0A0P5RD29 A0A0P5FJS2 A0A131XVY9 A0A162P4D0 B7P873 A0A0P5RY84 A0A0N8ANZ9 A0A2R5LBA6 A0A182STT6 A0A034WG87 L7MEL3 A0A0P4XBC1 A0A182Y2T4 A0A182J7F1 A0A224YS15 A0A2M4BNJ3 A0A0P5N6L6 A0A1I8M4H6 T1PEN9 A0A0L0C3N8
S4PYE3 A0A194R6B6 A0A0L7LCC2 A0A2P8Z8Y6 A0A2J7PFC0 A0A067QRT2 A0A1W4W452 A0A1Y1LZ61 A0A154P1C5 A0A2J7PFC9 E9IPB8 A0A0L7QT15 A0A195DRM3 A0A151I5S5 A0A158NAL2 A0A151WSY4 D6WJD0 F4X280 A0A1W4WFY2 E2A948 A0A2A3EGI2 A0A0C9RHL2 A0A0M9A650 E2BKR4 A0A1B6CVJ3 A0A088AW53 A0A026WAS7 A0A1B6C0Q8 K7IXK9 A0A3L8DGA9 A0A232ET99 A0A1B6KFK8 A0A195EVG9 A0A0J7N3E1 U4TZI2 J3JTQ0 N6U387 A0A0F6YT47 A0A1B6CE79 A0A182KP02 A0A182U1I1 Q7Q5Z7 A0A182I1F5 A0A182V7C4 B0WFM2 A0A182JUT7 Q17KI4 A0A224XG84 A0A1Q3F9Z2 A0A182PDU1 A0A023EWZ3 A0A182XP71 A0A0K8TQR3 E0VB34 A0A0V0G553 A0A1B6HJJ0 A0A069DTR3 A0A023EQ68 A0A0P4VW87 A0A182GGW4 A0A182N8A9 A0A182LYT4 A0A0P4WJT0 A0A182QT88 A0A1S3KCX6 A0A2T7NL92 A0A1S3KCD8 A0A182REY9 A0A1A9WTK3 E9HQF6 A0A0P5RD29 A0A0P5FJS2 A0A131XVY9 A0A162P4D0 B7P873 A0A0P5RY84 A0A0N8ANZ9 A0A2R5LBA6 A0A182STT6 A0A034WG87 L7MEL3 A0A0P4XBC1 A0A182Y2T4 A0A182J7F1 A0A224YS15 A0A2M4BNJ3 A0A0P5N6L6 A0A1I8M4H6 T1PEN9 A0A0L0C3N8
Ontologies
PANTHER
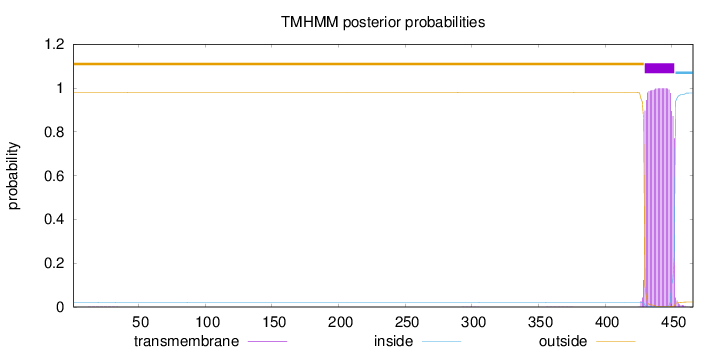
Topology
Length:
466
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.59879
Exp number, first 60 AAs:
0.00542
Total prob of N-in:
0.02132
outside
1 - 429
TMhelix
430 - 452
inside
453 - 466
Population Genetic Test Statistics
Pi
269.589227
Theta
212.854921
Tajima's D
1.97867
CLR
0.227974
CSRT
0.88075596220189
Interpretation
Uncertain
