Gene
KWMTBOMO13113
Pre Gene Modal
BGIBMGA000384
Annotation
PREDICTED:_uncharacterized_protein_LOC106105585_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.066 Mitochondrial Reliability : 1.358 Nuclear Reliability : 1.588
Sequence
CDS
ATGTCGGTAAAAGTATTTCCCGGCGAAAGTAAATACGAAACGACGCTACAAGAGAATTCATGGCCGGTATGGAACGAAGATTTCAAATTTCAATTAAAGCAAAAAGAAGCGAAGAAGTCGTCGGACAAAATTGACCTTCAGAATTTAGTATCCGGTCATTTTTTATCACTTACAATTTATGCTATATTAGAGCCGCCAAAACAAGAAGCCGACAAACGTAAAAACAGTAAGACAATAGATACTAAAAATGCAAAAGTAGGAGATCAAGAGGAACCCAAAGCTAAAACAGGTTTTCTGGAGAAAACATTCAGTAGTTTCAAAGCGACAAAAGCTGAAGCGATAGCACAAAAATCTATTTTGGAAAAACGACGGACCGTAGGGGCAGCCACTTGGAATTTTGACTCTAAATTATTTCAAAATGATTTGAAGAACGGTTTGATTGGAACTCCAGACATATGGAGGCCCATAAATGCGATTGCAAGTGGAATTACCGCAAGTGAAAAACGGGGTGAAAACACTAAAGGCCAACTGGAGGTGACCCTTTTGTACACGGCGAGCGAAGACGGTCTGAACGATGTGGTTCAGTTGACAGTAAACAGATTACGTTGCAGCGTCCACACAATGCAGGAACACGAGAAATATAAAGCACCTCTGTACCTGAAAGCGACTATATTGGAGGCGAACAAAGCCGAGTGCTTCTGGAAGAGCGACAGATTCGTCCCAACGATATCCGCCAGATGGGACCCTAAATCCGCGACAGTGAGGCTATCGGTTTTCAAAGCGAGCCTCGATCAAGTCAGCATTTATATAAGCTTAGGATGCAAAACGAAAATGGCGAAGAAGGAAATACTCGGAAAGGCGACAATACACGCTCAGTCTGCATATTCTGAAACGTGGACGCAATGTTTGAAGGAACCGGGCATACCGAAAACGTTCTGGGTCCAATTTCAATAA
Protein
MSVKVFPGESKYETTLQENSWPVWNEDFKFQLKQKEAKKSSDKIDLQNLVSGHFLSLTIYAILEPPKQEADKRKNSKTIDTKNAKVGDQEEPKAKTGFLEKTFSSFKATKAEAIAQKSILEKRRTVGAATWNFDSKLFQNDLKNGLIGTPDIWRPINAIASGITASEKRGENTKGQLEVTLLYTASEDGLNDVVQLTVNRLRCSVHTMQEHEKYKAPLYLKATILEANKAECFWKSDRFVPTISARWDPKSATVRLSVFKASLDQVSIYISLGCKTKMAKKEILGKATIHAQSAYSETWTQCLKEPGIPKTFWVQFQ
Summary
Uniprot
H9IT05
A0A212F9H2
A0A1E1WJ52
A0A194PXX6
A0A194R6B6
A0A0L7LBI4
+ More
A0A1W4W437 D6WJD6 A0A195ETG4 A0A151X202 A0A1Y1KE72 A0A154PCM8 F4W419 A0A195DVK0 A0A026WLN5 A0A195CLQ1 E9I9S8 A0A195B432 A0A0J7KXM9 A0A158NK10 E2BK79 A0A0L7QTA7 N6UD05 E2A339 A0A088AWA1 A0A2A3EGJ1 A0A0C9QQ88 A0A0M9A7E9 A0A0T6BH20 K7J1R6 A0A232FCC5 A0A336LHV0 A0A1J1IDV8 U5EPC7 A0A1B0CZH5 A0A0J9RU22 A0A0R1DXE9 B4QRB2 B4PGR4 A0A0Q5UAX5 B3NHA7 A0A0P8XUS1 B4J323 A0A3B0KGA9 A0A3B0KLH3 B3MB12 A0A2P8Z902
A0A1W4W437 D6WJD6 A0A195ETG4 A0A151X202 A0A1Y1KE72 A0A154PCM8 F4W419 A0A195DVK0 A0A026WLN5 A0A195CLQ1 E9I9S8 A0A195B432 A0A0J7KXM9 A0A158NK10 E2BK79 A0A0L7QTA7 N6UD05 E2A339 A0A088AWA1 A0A2A3EGJ1 A0A0C9QQ88 A0A0M9A7E9 A0A0T6BH20 K7J1R6 A0A232FCC5 A0A336LHV0 A0A1J1IDV8 U5EPC7 A0A1B0CZH5 A0A0J9RU22 A0A0R1DXE9 B4QRB2 B4PGR4 A0A0Q5UAX5 B3NHA7 A0A0P8XUS1 B4J323 A0A3B0KGA9 A0A3B0KLH3 B3MB12 A0A2P8Z902
Pubmed
EMBL
BABH01013124
AGBW02009593
OWR50363.1
GDQN01004042
JAT87012.1
KQ459586
+ More
KPI97878.1 KQ460883 KPJ11406.1 JTDY01001802 KOB72848.1 KQ971343 EFA03130.1 KQ981979 KYN31528.1 KQ982585 KYQ54250.1 GEZM01086089 JAV59782.1 KQ434870 KZC09602.1 GL887491 EGI71015.1 KQ980295 KYN16871.1 KK107153 QOIP01000012 EZA56955.1 RLU15791.1 KQ977642 KYN00999.1 GL761846 EFZ22686.1 KQ976625 KYM78969.1 LBMM01002268 KMQ95039.1 ADTU01018615 ADTU01018616 GL448770 EFN83876.1 KQ414755 KOC61804.1 APGK01028340 KB740686 KB631930 ENN79550.1 ERL87306.1 GL436272 EFN72142.1 KZ288263 PBC30339.1 GBYB01005854 JAG75621.1 KQ435727 KOX78006.1 LJIG01000463 KRT86477.1 NNAY01000427 OXU28476.1 UFQT01000007 SSX17456.1 CVRI01000047 CRK98403.1 GANO01004757 JAB55114.1 AJVK01020779 AJVK01020780 CM002912 KMY99238.1 CM000159 KRK01756.1 CM000363 EDX10242.1 KMY99237.1 EDW94303.1 CH954178 KQS43853.1 EDV51564.1 CH902618 KPU78477.1 CH916366 EDV96094.1 OUUW01000012 SPP87430.1 SPP87429.1 EDV40278.1 PYGN01000143 PSN52970.1
KPI97878.1 KQ460883 KPJ11406.1 JTDY01001802 KOB72848.1 KQ971343 EFA03130.1 KQ981979 KYN31528.1 KQ982585 KYQ54250.1 GEZM01086089 JAV59782.1 KQ434870 KZC09602.1 GL887491 EGI71015.1 KQ980295 KYN16871.1 KK107153 QOIP01000012 EZA56955.1 RLU15791.1 KQ977642 KYN00999.1 GL761846 EFZ22686.1 KQ976625 KYM78969.1 LBMM01002268 KMQ95039.1 ADTU01018615 ADTU01018616 GL448770 EFN83876.1 KQ414755 KOC61804.1 APGK01028340 KB740686 KB631930 ENN79550.1 ERL87306.1 GL436272 EFN72142.1 KZ288263 PBC30339.1 GBYB01005854 JAG75621.1 KQ435727 KOX78006.1 LJIG01000463 KRT86477.1 NNAY01000427 OXU28476.1 UFQT01000007 SSX17456.1 CVRI01000047 CRK98403.1 GANO01004757 JAB55114.1 AJVK01020779 AJVK01020780 CM002912 KMY99238.1 CM000159 KRK01756.1 CM000363 EDX10242.1 KMY99237.1 EDW94303.1 CH954178 KQS43853.1 EDV51564.1 CH902618 KPU78477.1 CH916366 EDV96094.1 OUUW01000012 SPP87430.1 SPP87429.1 EDV40278.1 PYGN01000143 PSN52970.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000037510
UP000192223
+ More
UP000007266 UP000078541 UP000075809 UP000076502 UP000007755 UP000078492 UP000053097 UP000279307 UP000078542 UP000078540 UP000036403 UP000005205 UP000008237 UP000053825 UP000019118 UP000030742 UP000000311 UP000005203 UP000242457 UP000053105 UP000002358 UP000215335 UP000183832 UP000092462 UP000002282 UP000000304 UP000008711 UP000007801 UP000001070 UP000268350 UP000245037
UP000007266 UP000078541 UP000075809 UP000076502 UP000007755 UP000078492 UP000053097 UP000279307 UP000078542 UP000078540 UP000036403 UP000005205 UP000008237 UP000053825 UP000019118 UP000030742 UP000000311 UP000005203 UP000242457 UP000053105 UP000002358 UP000215335 UP000183832 UP000092462 UP000002282 UP000000304 UP000008711 UP000007801 UP000001070 UP000268350 UP000245037
PRIDE
Gene 3D
ProteinModelPortal
H9IT05
A0A212F9H2
A0A1E1WJ52
A0A194PXX6
A0A194R6B6
A0A0L7LBI4
+ More
A0A1W4W437 D6WJD6 A0A195ETG4 A0A151X202 A0A1Y1KE72 A0A154PCM8 F4W419 A0A195DVK0 A0A026WLN5 A0A195CLQ1 E9I9S8 A0A195B432 A0A0J7KXM9 A0A158NK10 E2BK79 A0A0L7QTA7 N6UD05 E2A339 A0A088AWA1 A0A2A3EGJ1 A0A0C9QQ88 A0A0M9A7E9 A0A0T6BH20 K7J1R6 A0A232FCC5 A0A336LHV0 A0A1J1IDV8 U5EPC7 A0A1B0CZH5 A0A0J9RU22 A0A0R1DXE9 B4QRB2 B4PGR4 A0A0Q5UAX5 B3NHA7 A0A0P8XUS1 B4J323 A0A3B0KGA9 A0A3B0KLH3 B3MB12 A0A2P8Z902
A0A1W4W437 D6WJD6 A0A195ETG4 A0A151X202 A0A1Y1KE72 A0A154PCM8 F4W419 A0A195DVK0 A0A026WLN5 A0A195CLQ1 E9I9S8 A0A195B432 A0A0J7KXM9 A0A158NK10 E2BK79 A0A0L7QTA7 N6UD05 E2A339 A0A088AWA1 A0A2A3EGJ1 A0A0C9QQ88 A0A0M9A7E9 A0A0T6BH20 K7J1R6 A0A232FCC5 A0A336LHV0 A0A1J1IDV8 U5EPC7 A0A1B0CZH5 A0A0J9RU22 A0A0R1DXE9 B4QRB2 B4PGR4 A0A0Q5UAX5 B3NHA7 A0A0P8XUS1 B4J323 A0A3B0KGA9 A0A3B0KLH3 B3MB12 A0A2P8Z902
Ontologies
GO
PANTHER
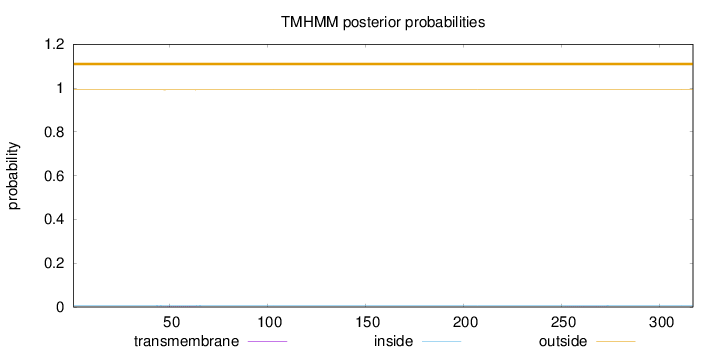
Topology
Length:
317
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0331
Exp number, first 60 AAs:
0.02284
Total prob of N-in:
0.00759
outside
1 - 317
Population Genetic Test Statistics
Pi
208.469181
Theta
161.797033
Tajima's D
0.819429
CLR
0.084962
CSRT
0.6037198140093
Interpretation
Uncertain
