Gene
KWMTBOMO13112
Pre Gene Modal
BGIBMGA000385
Annotation
PREDICTED:_elastase-1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.15
Sequence
CDS
ATGTGGATACGACCTGCAATCATTTTAAATTTAATCACCATCGAATTTTCTTTAACTAATTATATAGAAAACGATCCGATATCGACCGAGGGGCATGCGAATTTATTTGACATGAAAACATCTAAAAAGTATCAAAATATTAGATTGATGGAACCAAAAAAAGGAATTCATTTAAATCGACCTTCATATTCGCCGTGGACTCGATGGAGTGTATGCCAAGAAGGTCGTAAGACCCGGAGGCGACATTGTGTACTTAAAAGAGTGTGTGGGGACTCCTTACGCGTTCAAGTTTCAAGATGTAGACAACGCAACAGGGGTCGAAAGAAGATGACACAAAACACAAAGTTCGGAAATTACATGATTATACCTCAAAGACGTCAGTTAGATTCCGAAGATGTGCAGAAACGATTCCGTGGATTCTCGCAGTGGAGCTCATGGACTAATTGTTCTCGCAAATGTACAACTGTAAGAAGAAGGCGATGCATGAAACGGTCAATTTGCGGTCGTAAAATTGTAAAGCACTCGGCGTACTGCTATTTGGAAGGGAGTTTCTGTCACACGTGGATACGAAACAGAATGCAGAAGCGGCAGGACCCAGGGCGTGCAGTATTACAAGCCATGCCTCCACCGGCTCCGGCGATACATCACGAAGATTACCGAATGGCTCCGACTGCCCACTCCTGTGGTCGACTAGGACGGTATCGAGGCTCTGTAGCCGCGAGGTACAGGGGACGTATGAGAGACATGATCCGGATCATCGGAGGAAGGCCGGCGCCGCCTGGTAAATGGCCGTGGCAGGTTGCCGTTCTAAATCGATTTAAGGAGGCGTTTTGTGGCGGGACCCTCGTTTCGCTTCGCTGGGTGGTGACAGCAGCTCATTGCGTGCGGCGACGGCTTTACGTGCGCTTGGGTGAGCACGACCTGCTCCTGCGGAACTACGGGGAACTGGAGATGAAGGTCACCGAAGCAGTCGTACATCCTCGATACGACCCTGACACGGTCATCAACGATGTCGCCATGCTCCGATTACCGACGTCCGCCCGCCCCGACCTTGGTCACGGGATAGCTTGCCTTCCGAATCCACACCAGTCTCTCCCTCCGCATACATCCTGCATAATACTCGGGTGGGGTAAGAAGCGGCCCACAGATGTCCACGGTACGAGGGTGTTGCACGAAGCGCAGGTTTCGACAATACAGCAAGGCGTATGTCGTCGCTCGTACTGGCAATACGCCATCACAGACGACATGGTGTGTGCGGGACGCGGGCGGCGTGATTCGTGCGCAGGGGACTCCGGCGGACCGCTGCTTTGTCGGGACCGCTCCAATAGATACGTCTTACAGGGCATTACCAGTTTCGGAGACGGATGCGGGAAGCGTGGGAAGTACGGCATATACACGAGAACTGCTGGTTACGTCACTTGGATTAAAGACGTAATGAATAGCAAATACTTCGATTGA
Protein
MWIRPAIILNLITIEFSLTNYIENDPISTEGHANLFDMKTSKKYQNIRLMEPKKGIHLNRPSYSPWTRWSVCQEGRKTRRRHCVLKRVCGDSLRVQVSRCRQRNRGRKKMTQNTKFGNYMIIPQRRQLDSEDVQKRFRGFSQWSSWTNCSRKCTTVRRRRCMKRSICGRKIVKHSAYCYLEGSFCHTWIRNRMQKRQDPGRAVLQAMPPPAPAIHHEDYRMAPTAHSCGRLGRYRGSVAARYRGRMRDMIRIIGGRPAPPGKWPWQVAVLNRFKEAFCGGTLVSLRWVVTAAHCVRRRLYVRLGEHDLLLRNYGELEMKVTEAVVHPRYDPDTVINDVAMLRLPTSARPDLGHGIACLPNPHQSLPPHTSCIILGWGKKRPTDVHGTRVLHEAQVSTIQQGVCRRSYWQYAITDDMVCAGRGRRDSCAGDSGGPLLCRDRSNRYVLQGITSFGDGCGKRGKYGIYTRTAGYVTWIKDVMNSKYFD
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A194PX27
A0A194R126
A0A088AWA3
A0A154PDX0
A0A195CJU6
A0A2A3EH64
+ More
A0A3L8DTA9 E2A336 A0A026VUU7 A0A195DVG5 A0A0C9R8P9 A0A151X1H3 A0A0C9RPV9 A0A158NK12 A0A195ETP6 E2BK82 E9I9S7 F4W417 A0A1B6F2I2 A0A195B3X0 A0A069DU95 A0A0L7QTK2 A0A1V1FT88 A0A023F1G6 A0A0P4VMX2 K7J0S8 A0A0P5D951 A0A0P5HHT6 A0A0P5DHC5 A0A0P5C4N1 A0A0P5C4V5 A0A0P5G7K9 A0A0P5EL82 A0A0P6GJY0 A0A0P5NCM9 A0A0P5KI61 A0A0P5DGZ1 A0A0N1ITX2 A0A0N8C036 B3MB11 A0A182GLQ6 Q16QB1 A0A0P5IZR6 A0A1A9Y0T8 A0A0P6HCS1 A0A1W4UUK0 A0A1A9V3Q2 A0A1B0G8G9 A0A1B0AI58 A0A1A9WTK5 A0A0P5VFC5 A8JNS0 A0A0P5N4V8 A0A0P5ZV50 B3NHA6 A0A0J9UJV3 Q2M0M7 B4GRF1 B4PGR2 T1J342 A0A0P5CXR2 A0A336LIL7 A0A2R7WNQ4 A0A0P5HU33 B4LGN8 A0A3B0KU27 A0A0P5MUA5 B4J324 B4L078 A0A1J1IDQ2 A0A1J1IH93 A0A1B0B5K3 A0A0P5TFI5 A0A084VW61 T1JFB1 V5GYX6 D6WJD5
A0A3L8DTA9 E2A336 A0A026VUU7 A0A195DVG5 A0A0C9R8P9 A0A151X1H3 A0A0C9RPV9 A0A158NK12 A0A195ETP6 E2BK82 E9I9S7 F4W417 A0A1B6F2I2 A0A195B3X0 A0A069DU95 A0A0L7QTK2 A0A1V1FT88 A0A023F1G6 A0A0P4VMX2 K7J0S8 A0A0P5D951 A0A0P5HHT6 A0A0P5DHC5 A0A0P5C4N1 A0A0P5C4V5 A0A0P5G7K9 A0A0P5EL82 A0A0P6GJY0 A0A0P5NCM9 A0A0P5KI61 A0A0P5DGZ1 A0A0N1ITX2 A0A0N8C036 B3MB11 A0A182GLQ6 Q16QB1 A0A0P5IZR6 A0A1A9Y0T8 A0A0P6HCS1 A0A1W4UUK0 A0A1A9V3Q2 A0A1B0G8G9 A0A1B0AI58 A0A1A9WTK5 A0A0P5VFC5 A8JNS0 A0A0P5N4V8 A0A0P5ZV50 B3NHA6 A0A0J9UJV3 Q2M0M7 B4GRF1 B4PGR2 T1J342 A0A0P5CXR2 A0A336LIL7 A0A2R7WNQ4 A0A0P5HU33 B4LGN8 A0A3B0KU27 A0A0P5MUA5 B4J324 B4L078 A0A1J1IDQ2 A0A1J1IH93 A0A1B0B5K3 A0A0P5TFI5 A0A084VW61 T1JFB1 V5GYX6 D6WJD5
Pubmed
26354079
30249741
20798317
24508170
21347285
21282665
+ More
21719571 26334808 28410430 25474469 27129103 20075255 17994087 18057021 26483478 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 23185243 17550304 24438588 18362917 19820115
21719571 26334808 28410430 25474469 27129103 20075255 17994087 18057021 26483478 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 23185243 17550304 24438588 18362917 19820115
EMBL
KQ459586
KPI97877.1
KQ460883
KPJ11407.1
KQ434870
KZC09604.1
+ More
KQ977642 KYN01001.1 KZ288263 PBC30341.1 QOIP01000005 RLU22998.1 GL436272 EFN72139.1 KK107847 EZA47450.1 KQ980295 KYN16873.1 GBYB01009222 JAG78989.1 KQ982585 KYQ54247.1 GBYB01009219 GBYB01009220 GBYB01009221 JAG78986.1 JAG78987.1 JAG78988.1 ADTU01018618 ADTU01018619 KQ981979 KYN31531.1 GL448770 EFN83879.1 GL761846 EFZ22722.1 GL887491 EGI71013.1 GECZ01025374 JAS44395.1 KQ976625 KYM78967.1 GBGD01001379 JAC87510.1 KQ414755 KOC61806.1 FX985322 BAX07335.1 GBBI01003878 JAC14834.1 GDKW01002911 JAI53684.1 GDIP01175566 JAJ47836.1 GDIQ01249890 JAK01835.1 GDIP01156186 JAJ67216.1 GDIP01175567 JAJ47835.1 GDIP01175568 JAJ47834.1 GDIQ01245151 JAK06574.1 GDIP01146579 JAJ76823.1 GDIQ01034464 JAN60273.1 GDIQ01147133 JAL04593.1 GDIQ01185007 GDIP01073193 JAK66718.1 JAM30522.1 GDIP01156397 JAJ67005.1 KQ435727 KOX78002.1 GDIQ01127971 JAL23755.1 CH902618 EDV40277.1 KPU78476.1 JXUM01072683 JXUM01072684 JXUM01072685 JXUM01072686 JXUM01072687 JXUM01072688 KQ562734 KXJ75235.1 CH477755 EAT36577.1 GDIQ01207151 JAK44574.1 GDIQ01021376 JAN73361.1 CCAG010000255 GDIP01100729 JAM02986.1 AE014296 ABW08531.1 ABW08532.2 GDIQ01151127 JAL00599.1 GDIP01042540 JAM61175.1 CH954178 EDV51563.2 CM002912 KMY99235.1 CH379069 EAL30903.2 KRT07752.1 CH479188 EDW40336.1 CM000159 EDW94301.2 KRK01755.1 JH431820 GDIP01164132 JAJ59270.1 UFQT01000007 SSX17455.1 KK855062 PTY20620.1 GDIQ01228089 JAK23636.1 CH940647 EDW69476.1 OUUW01000012 SPP87428.1 GDIQ01151126 JAL00600.1 CH916366 EDV96095.1 CH933809 EDW18024.1 KRG05906.1 KRG05907.1 CVRI01000047 CRK98407.1 CRK98406.1 JXJN01008760 JXJN01008761 GDIP01132792 JAL70922.1 ATLV01017482 ATLV01017483 ATLV01017484 ATLV01017485 ATLV01017486 ATLV01017487 ATLV01017488 ATLV01017489 KE525170 KFB42205.1 JH432147 GALX01002808 JAB65658.1 KQ971343 EFA04569.1
KQ977642 KYN01001.1 KZ288263 PBC30341.1 QOIP01000005 RLU22998.1 GL436272 EFN72139.1 KK107847 EZA47450.1 KQ980295 KYN16873.1 GBYB01009222 JAG78989.1 KQ982585 KYQ54247.1 GBYB01009219 GBYB01009220 GBYB01009221 JAG78986.1 JAG78987.1 JAG78988.1 ADTU01018618 ADTU01018619 KQ981979 KYN31531.1 GL448770 EFN83879.1 GL761846 EFZ22722.1 GL887491 EGI71013.1 GECZ01025374 JAS44395.1 KQ976625 KYM78967.1 GBGD01001379 JAC87510.1 KQ414755 KOC61806.1 FX985322 BAX07335.1 GBBI01003878 JAC14834.1 GDKW01002911 JAI53684.1 GDIP01175566 JAJ47836.1 GDIQ01249890 JAK01835.1 GDIP01156186 JAJ67216.1 GDIP01175567 JAJ47835.1 GDIP01175568 JAJ47834.1 GDIQ01245151 JAK06574.1 GDIP01146579 JAJ76823.1 GDIQ01034464 JAN60273.1 GDIQ01147133 JAL04593.1 GDIQ01185007 GDIP01073193 JAK66718.1 JAM30522.1 GDIP01156397 JAJ67005.1 KQ435727 KOX78002.1 GDIQ01127971 JAL23755.1 CH902618 EDV40277.1 KPU78476.1 JXUM01072683 JXUM01072684 JXUM01072685 JXUM01072686 JXUM01072687 JXUM01072688 KQ562734 KXJ75235.1 CH477755 EAT36577.1 GDIQ01207151 JAK44574.1 GDIQ01021376 JAN73361.1 CCAG010000255 GDIP01100729 JAM02986.1 AE014296 ABW08531.1 ABW08532.2 GDIQ01151127 JAL00599.1 GDIP01042540 JAM61175.1 CH954178 EDV51563.2 CM002912 KMY99235.1 CH379069 EAL30903.2 KRT07752.1 CH479188 EDW40336.1 CM000159 EDW94301.2 KRK01755.1 JH431820 GDIP01164132 JAJ59270.1 UFQT01000007 SSX17455.1 KK855062 PTY20620.1 GDIQ01228089 JAK23636.1 CH940647 EDW69476.1 OUUW01000012 SPP87428.1 GDIQ01151126 JAL00600.1 CH916366 EDV96095.1 CH933809 EDW18024.1 KRG05906.1 KRG05907.1 CVRI01000047 CRK98407.1 CRK98406.1 JXJN01008760 JXJN01008761 GDIP01132792 JAL70922.1 ATLV01017482 ATLV01017483 ATLV01017484 ATLV01017485 ATLV01017486 ATLV01017487 ATLV01017488 ATLV01017489 KE525170 KFB42205.1 JH432147 GALX01002808 JAB65658.1 KQ971343 EFA04569.1
Proteomes
UP000053268
UP000053240
UP000005203
UP000076502
UP000078542
UP000242457
+ More
UP000279307 UP000000311 UP000053097 UP000078492 UP000075809 UP000005205 UP000078541 UP000008237 UP000007755 UP000078540 UP000053825 UP000002358 UP000053105 UP000007801 UP000069940 UP000249989 UP000008820 UP000092443 UP000192221 UP000078200 UP000092444 UP000092445 UP000091820 UP000000803 UP000008711 UP000001819 UP000008744 UP000002282 UP000008792 UP000268350 UP000001070 UP000009192 UP000183832 UP000092460 UP000030765 UP000007266
UP000279307 UP000000311 UP000053097 UP000078492 UP000075809 UP000005205 UP000078541 UP000008237 UP000007755 UP000078540 UP000053825 UP000002358 UP000053105 UP000007801 UP000069940 UP000249989 UP000008820 UP000092443 UP000192221 UP000078200 UP000092444 UP000092445 UP000091820 UP000000803 UP000008711 UP000001819 UP000008744 UP000002282 UP000008792 UP000268350 UP000001070 UP000009192 UP000183832 UP000092460 UP000030765 UP000007266
Interpro
CDD
ProteinModelPortal
A0A194PX27
A0A194R126
A0A088AWA3
A0A154PDX0
A0A195CJU6
A0A2A3EH64
+ More
A0A3L8DTA9 E2A336 A0A026VUU7 A0A195DVG5 A0A0C9R8P9 A0A151X1H3 A0A0C9RPV9 A0A158NK12 A0A195ETP6 E2BK82 E9I9S7 F4W417 A0A1B6F2I2 A0A195B3X0 A0A069DU95 A0A0L7QTK2 A0A1V1FT88 A0A023F1G6 A0A0P4VMX2 K7J0S8 A0A0P5D951 A0A0P5HHT6 A0A0P5DHC5 A0A0P5C4N1 A0A0P5C4V5 A0A0P5G7K9 A0A0P5EL82 A0A0P6GJY0 A0A0P5NCM9 A0A0P5KI61 A0A0P5DGZ1 A0A0N1ITX2 A0A0N8C036 B3MB11 A0A182GLQ6 Q16QB1 A0A0P5IZR6 A0A1A9Y0T8 A0A0P6HCS1 A0A1W4UUK0 A0A1A9V3Q2 A0A1B0G8G9 A0A1B0AI58 A0A1A9WTK5 A0A0P5VFC5 A8JNS0 A0A0P5N4V8 A0A0P5ZV50 B3NHA6 A0A0J9UJV3 Q2M0M7 B4GRF1 B4PGR2 T1J342 A0A0P5CXR2 A0A336LIL7 A0A2R7WNQ4 A0A0P5HU33 B4LGN8 A0A3B0KU27 A0A0P5MUA5 B4J324 B4L078 A0A1J1IDQ2 A0A1J1IH93 A0A1B0B5K3 A0A0P5TFI5 A0A084VW61 T1JFB1 V5GYX6 D6WJD5
A0A3L8DTA9 E2A336 A0A026VUU7 A0A195DVG5 A0A0C9R8P9 A0A151X1H3 A0A0C9RPV9 A0A158NK12 A0A195ETP6 E2BK82 E9I9S7 F4W417 A0A1B6F2I2 A0A195B3X0 A0A069DU95 A0A0L7QTK2 A0A1V1FT88 A0A023F1G6 A0A0P4VMX2 K7J0S8 A0A0P5D951 A0A0P5HHT6 A0A0P5DHC5 A0A0P5C4N1 A0A0P5C4V5 A0A0P5G7K9 A0A0P5EL82 A0A0P6GJY0 A0A0P5NCM9 A0A0P5KI61 A0A0P5DGZ1 A0A0N1ITX2 A0A0N8C036 B3MB11 A0A182GLQ6 Q16QB1 A0A0P5IZR6 A0A1A9Y0T8 A0A0P6HCS1 A0A1W4UUK0 A0A1A9V3Q2 A0A1B0G8G9 A0A1B0AI58 A0A1A9WTK5 A0A0P5VFC5 A8JNS0 A0A0P5N4V8 A0A0P5ZV50 B3NHA6 A0A0J9UJV3 Q2M0M7 B4GRF1 B4PGR2 T1J342 A0A0P5CXR2 A0A336LIL7 A0A2R7WNQ4 A0A0P5HU33 B4LGN8 A0A3B0KU27 A0A0P5MUA5 B4J324 B4L078 A0A1J1IDQ2 A0A1J1IH93 A0A1B0B5K3 A0A0P5TFI5 A0A084VW61 T1JFB1 V5GYX6 D6WJD5
PDB
5UGG
E-value=6.50721e-37,
Score=387
Ontologies
GO
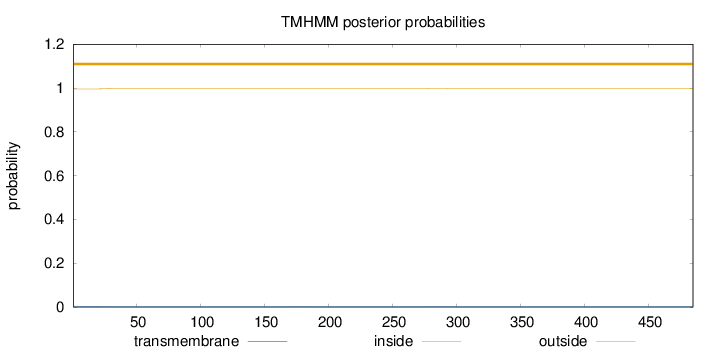
Topology
Length:
485
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0432700000000001
Exp number, first 60 AAs:
0.03557
Total prob of N-in:
0.00421
outside
1 - 485
Population Genetic Test Statistics
Pi
219.133147
Theta
176.512854
Tajima's D
0.410902
CLR
0.420533
CSRT
0.484125793710315
Interpretation
Uncertain
