Gene
KWMTBOMO13108
Annotation
PREDICTED:_glutamine_synthetase_2_cytoplasmic-like_[Plutella_xylostella]
Full name
Glutamine synthetase
+ More
Glutamine synthetase 2 cytoplasmic
Glutamine synthetase 2 cytoplasmic
Alternative Name
Glutamate--ammonia ligase 2
Location in the cell
Extracellular Reliability : 1.054
Sequence
CDS
ATGTTACCGGTGTTTTTTTCTTTTCTTTCAACAGCGTCAAATCACCGGAAAAACTGTACGATAATATGCGAGAAGGGTGAAGTGGAGGAGCCCTGGTTTGGCTTTAATCAAGAATTCATCTTGACATCGTCTGATGGACGTCCCCTAGGTTGGCCCGTTGGTGGATTTCCCGCGCCTCCCGGGCCTTATTACTGCGCCATAGGGTCGGATAAAATAGTTGCGAGAGATCTTATGGAAGCCTTTTACAGGTGCTGTTTATACGCTGGAGTTCAGTTAAATGGAATAAATCCGGGAACGGTGCCCTCCCAGTGGAACTTCCAGGTCGGTCCAAGTCCTGGTATTCGTGGAGCGGATGATTTATGGATGGCTCGGTACATTTTATCACGACTCGCCGAGGAGTACGGCACGGTGGCCTCCTTCGAACCACAGCCCACAGCGGATTGGCCGGGAAACGGTACCTTTGTGTACTTTTCCACCAAGGACATGAGAGAAGATGACGGCATATTAGTGATCGAGCGGGCTATAGATAAGTTGGCGCGTCGACACGGTGCCCACATCAATGAGTACGACGCTAACAATGGGGCTGACAACGTGCGAAGGCTCGTCGGAAAAAATGGAATGCCACACATCAGGGATTTCACTGCAGGTGTGGCTAATCGGAGTTGTTGCGTCAGGATACCTCGGCAAGTTTCCGAGGACAAGCGGGGCTATCTAGAAGATAGGCGACCCGCAGCTAACGCAGACCCGTACCGCATCATATCAATTATGCTCAAAACTTGTATTTTTGATGAGTGA
Protein
MLPVFFSFLSTASNHRKNCTIICEKGEVEEPWFGFNQEFILTSSDGRPLGWPVGGFPAPPGPYYCAIGSDKIVARDLMEAFYRCCLYAGVQLNGINPGTVPSQWNFQVGPSPGIRGADDLWMARYILSRLAEEYGTVASFEPQPTADWPGNGTFVYFSTKDMREDDGILVIERAIDKLARRHGAHINEYDANNGADNVRRLVGKNGMPHIRDFTAGVANRSCCVRIPRQVSEDKRGYLEDRRPAANADPYRIISIMLKTCIFDE
Summary
Catalytic Activity
ATP + L-glutamate + NH4(+) = ADP + H(+) + L-glutamine + phosphate
Subunit
Homooctamer.
Similarity
Belongs to the glutamine synthetase family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Ligase
Nucleotide-binding
Reference proteome
Feature
chain Glutamine synthetase 2 cytoplasmic
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A3S2PC21
A0A194R0T4
A0A194Q394
A0A0L7KZK1
A0A2H1VSF8
A0A067R5C5
+ More
A0A068TK10 A0A2J7RJB5 A0A0P4VPJ5 R4G5A5 Q5PRD7 A0A1Y1KMB3 V5HHJ9 A0A1Z5LAY3 A0A1J1HER3 A0A293LB34 A0A154P8H2 E0VED6 A0A151JWT9 A0A1B6IKT6 F4WK81 A0A1B6G578 A0A1B6FLE3 A0A158NIH3 A0A195BGP1 T1IUE5 A0A0N7ZBI5 J3JTU0 A0A151XJR8 A0A195C1J9 E2BUV7 A0A171A231 A0A182PA28 A0A2H8U045 M4HZ20 A0A1I9WLM6 A0A023GLZ5 T1H9Q5 G3MQD4 A0A1W4WLS5 H9N535 A0A182Q4C3 A0A165RB61 D6WFH1 A0A087UK01 A0A182U6F2 A0A182XIC8 A0A182LFG0 Q7QBX9 A0A182V4E1 F5HK76 A0A182HTB3 A0A1E1X7X7 A0A088AIM3 A0A0G3VIA2 A0A131YQE5 P20478-2 A0A1D1WAJ9 A0A023FZD1 R4WR46 A0A0A1XT45 U5EZ20 A0A2W1BMY2 A0A0A1WVL5 B4L4A8 A0A182JIW4 A0A0Q9WMW7 A0A2H1VZ23 A0A023FS93 I4DJ32 J9JML2 A0A182MEI4 L7M7P1 A0A224YQG5 B4R2U1 A0A088A2A8 V5G3M6 V9IDX6 A0A0G2QRP6 A0A084VJR5 A0A023F924 A0A1B6IB03 A0A2A3E9Y7 A0A2A4J4U6 M9V250 A0A131XHK3 A0A3R7LSZ4 A0A194QUW7 C0KJQ3 A0A1A9WZ89 A0A1I8NDX3 E2ATW3 B4JIW8 A0A182N4N2 A0A194PS35 H2BL56 K7IR04 X2JDA5 A0A182K3Y7 A0A2C9K0W2
A0A068TK10 A0A2J7RJB5 A0A0P4VPJ5 R4G5A5 Q5PRD7 A0A1Y1KMB3 V5HHJ9 A0A1Z5LAY3 A0A1J1HER3 A0A293LB34 A0A154P8H2 E0VED6 A0A151JWT9 A0A1B6IKT6 F4WK81 A0A1B6G578 A0A1B6FLE3 A0A158NIH3 A0A195BGP1 T1IUE5 A0A0N7ZBI5 J3JTU0 A0A151XJR8 A0A195C1J9 E2BUV7 A0A171A231 A0A182PA28 A0A2H8U045 M4HZ20 A0A1I9WLM6 A0A023GLZ5 T1H9Q5 G3MQD4 A0A1W4WLS5 H9N535 A0A182Q4C3 A0A165RB61 D6WFH1 A0A087UK01 A0A182U6F2 A0A182XIC8 A0A182LFG0 Q7QBX9 A0A182V4E1 F5HK76 A0A182HTB3 A0A1E1X7X7 A0A088AIM3 A0A0G3VIA2 A0A131YQE5 P20478-2 A0A1D1WAJ9 A0A023FZD1 R4WR46 A0A0A1XT45 U5EZ20 A0A2W1BMY2 A0A0A1WVL5 B4L4A8 A0A182JIW4 A0A0Q9WMW7 A0A2H1VZ23 A0A023FS93 I4DJ32 J9JML2 A0A182MEI4 L7M7P1 A0A224YQG5 B4R2U1 A0A088A2A8 V5G3M6 V9IDX6 A0A0G2QRP6 A0A084VJR5 A0A023F924 A0A1B6IB03 A0A2A3E9Y7 A0A2A4J4U6 M9V250 A0A131XHK3 A0A3R7LSZ4 A0A194QUW7 C0KJQ3 A0A1A9WZ89 A0A1I8NDX3 E2ATW3 B4JIW8 A0A182N4N2 A0A194PS35 H2BL56 K7IR04 X2JDA5 A0A182K3Y7 A0A2C9K0W2
EC Number
6.3.1.2
Pubmed
26354079
26227816
24845553
29403074
27129103
28004739
+ More
25765539 28528879 20566863 21719571 21347285 22516182 20798317 27538518 22216098 18362917 19820115 20966253 12364791 14747013 17210077 28503490 26830274 1969491 10731132 12537572 12537569 27649274 23691247 25830018 28756777 17994087 22651552 25576852 28797301 24438588 25474469 28049606 25315136 20075255 12537568 12537573 12537574 16110336 17569856 17569867 15562597
25765539 28528879 20566863 21719571 21347285 22516182 20798317 27538518 22216098 18362917 19820115 20966253 12364791 14747013 17210077 28503490 26830274 1969491 10731132 12537572 12537569 27649274 23691247 25830018 28756777 17994087 22651552 25576852 28797301 24438588 25474469 28049606 25315136 20075255 12537568 12537573 12537574 16110336 17569856 17569867 15562597
EMBL
RSAL01000111
RVE47082.1
KQ460883
KPJ11408.1
KQ459586
KPI97875.1
+ More
JTDY01004164 KOB68506.1 ODYU01004166 SOQ43749.1 KK852685 KDR18484.1 HG965789 PYGN01000753 CDO39385.1 PSN41298.1 NEVH01002994 PNF40921.1 GDKW01001673 JAI54922.1 GAHY01000111 JAA77399.1 BC086702 AAH86702.1 GEZM01081263 GEZM01081262 JAV61728.1 GANP01009731 JAB74737.1 GFJQ02002869 JAW04101.1 CVRI01000001 CRK86464.1 GFWV01000251 MAA24981.1 KQ434844 KZC08141.1 DS235090 EEB11742.1 KQ981622 KYN39215.1 GECU01020177 JAS87529.1 GL888197 EGI65384.1 GECZ01012197 JAS57572.1 GECZ01018741 JAS51028.1 ADTU01016600 ADTU01016601 ADTU01016602 ADTU01016603 ADTU01016604 ADTU01016605 ADTU01016606 ADTU01016607 KQ976491 KYM83318.1 AFFK01019467 GDRN01082973 JAI61744.1 BT126649 AEE61612.1 KQ982068 KYQ60545.1 KQ978379 KYM94490.1 GL450772 EFN80494.1 GEMB01001576 JAS01583.1 GFXV01007809 GFXV01008140 MBW19614.1 MBW19945.1 JQ670884 AFV39703.1 KU932410 APA34046.1 GBBM01001280 JAC34138.1 ACPB03017902 ACPB03017903 JO844085 AEO35702.1 JF738076 AFD52981.1 AXCN02000052 KR028426 AML23852.1 KQ971327 EEZ99821.1 KK120178 KFM77690.1 AAAB01008859 EAA08219.5 EGK96917.1 APCN01000435 GFAC01004037 JAT95151.1 KP657638 RSAL01000006 AKL78863.1 RVE54315.1 GEDV01008241 JAP80316.1 X52759 AE014298 AY060701 BDGG01000019 GAV08884.1 GBBL01001232 JAC26088.1 AK417132 BAN20347.1 GBXI01000554 JAD13738.1 GANO01001583 JAB58288.1 KZ150069 PZC74080.1 GBXI01011834 JAD02458.1 CH933810 EDW07386.1 KRF94024.1 ODYU01005170 SOQ45772.1 GBBK01000869 JAC23613.1 AK401300 BAM17922.1 ABLF02031791 AXCM01011565 GACK01004713 JAA60321.1 GFPF01008029 MAA19175.1 CM000366 EDX17640.1 GALX01003841 JAB64625.1 JR039080 AEY58519.1 KC344400 AGY46258.1 ATLV01013858 KE524905 KFB38209.1 GBBI01001238 JAC17474.1 GECU01023633 JAS84073.1 KZ288311 PBC28530.1 NWSH01003020 PCG67105.1 KC445137 AGJ70738.1 GEFH01002032 JAP66549.1 QCYY01003402 ROT63652.1 KQ461108 KPJ09262.1 FJ612107 ACM89168.2 GL442742 EFN63125.1 CH916370 EDV99532.1 KQ459594 KPI96122.1 JF694444 AEX31557.1 AHN59591.1
JTDY01004164 KOB68506.1 ODYU01004166 SOQ43749.1 KK852685 KDR18484.1 HG965789 PYGN01000753 CDO39385.1 PSN41298.1 NEVH01002994 PNF40921.1 GDKW01001673 JAI54922.1 GAHY01000111 JAA77399.1 BC086702 AAH86702.1 GEZM01081263 GEZM01081262 JAV61728.1 GANP01009731 JAB74737.1 GFJQ02002869 JAW04101.1 CVRI01000001 CRK86464.1 GFWV01000251 MAA24981.1 KQ434844 KZC08141.1 DS235090 EEB11742.1 KQ981622 KYN39215.1 GECU01020177 JAS87529.1 GL888197 EGI65384.1 GECZ01012197 JAS57572.1 GECZ01018741 JAS51028.1 ADTU01016600 ADTU01016601 ADTU01016602 ADTU01016603 ADTU01016604 ADTU01016605 ADTU01016606 ADTU01016607 KQ976491 KYM83318.1 AFFK01019467 GDRN01082973 JAI61744.1 BT126649 AEE61612.1 KQ982068 KYQ60545.1 KQ978379 KYM94490.1 GL450772 EFN80494.1 GEMB01001576 JAS01583.1 GFXV01007809 GFXV01008140 MBW19614.1 MBW19945.1 JQ670884 AFV39703.1 KU932410 APA34046.1 GBBM01001280 JAC34138.1 ACPB03017902 ACPB03017903 JO844085 AEO35702.1 JF738076 AFD52981.1 AXCN02000052 KR028426 AML23852.1 KQ971327 EEZ99821.1 KK120178 KFM77690.1 AAAB01008859 EAA08219.5 EGK96917.1 APCN01000435 GFAC01004037 JAT95151.1 KP657638 RSAL01000006 AKL78863.1 RVE54315.1 GEDV01008241 JAP80316.1 X52759 AE014298 AY060701 BDGG01000019 GAV08884.1 GBBL01001232 JAC26088.1 AK417132 BAN20347.1 GBXI01000554 JAD13738.1 GANO01001583 JAB58288.1 KZ150069 PZC74080.1 GBXI01011834 JAD02458.1 CH933810 EDW07386.1 KRF94024.1 ODYU01005170 SOQ45772.1 GBBK01000869 JAC23613.1 AK401300 BAM17922.1 ABLF02031791 AXCM01011565 GACK01004713 JAA60321.1 GFPF01008029 MAA19175.1 CM000366 EDX17640.1 GALX01003841 JAB64625.1 JR039080 AEY58519.1 KC344400 AGY46258.1 ATLV01013858 KE524905 KFB38209.1 GBBI01001238 JAC17474.1 GECU01023633 JAS84073.1 KZ288311 PBC28530.1 NWSH01003020 PCG67105.1 KC445137 AGJ70738.1 GEFH01002032 JAP66549.1 QCYY01003402 ROT63652.1 KQ461108 KPJ09262.1 FJ612107 ACM89168.2 GL442742 EFN63125.1 CH916370 EDV99532.1 KQ459594 KPI96122.1 JF694444 AEX31557.1 AHN59591.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000037510
UP000027135
UP000245037
+ More
UP000235965 UP000183832 UP000076502 UP000009046 UP000078541 UP000007755 UP000005205 UP000078540 UP000075809 UP000078542 UP000008237 UP000075885 UP000015103 UP000192223 UP000075886 UP000007266 UP000054359 UP000075902 UP000076407 UP000075882 UP000007062 UP000075903 UP000075840 UP000005203 UP000000803 UP000186922 UP000009192 UP000075880 UP000007819 UP000075883 UP000000304 UP000030765 UP000242457 UP000218220 UP000283509 UP000091820 UP000095301 UP000000311 UP000001070 UP000075884 UP000002358 UP000075881 UP000076420
UP000235965 UP000183832 UP000076502 UP000009046 UP000078541 UP000007755 UP000005205 UP000078540 UP000075809 UP000078542 UP000008237 UP000075885 UP000015103 UP000192223 UP000075886 UP000007266 UP000054359 UP000075902 UP000076407 UP000075882 UP000007062 UP000075903 UP000075840 UP000005203 UP000000803 UP000186922 UP000009192 UP000075880 UP000007819 UP000075883 UP000000304 UP000030765 UP000242457 UP000218220 UP000283509 UP000091820 UP000095301 UP000000311 UP000001070 UP000075884 UP000002358 UP000075881 UP000076420
Interpro
Gene 3D
ProteinModelPortal
A0A3S2PC21
A0A194R0T4
A0A194Q394
A0A0L7KZK1
A0A2H1VSF8
A0A067R5C5
+ More
A0A068TK10 A0A2J7RJB5 A0A0P4VPJ5 R4G5A5 Q5PRD7 A0A1Y1KMB3 V5HHJ9 A0A1Z5LAY3 A0A1J1HER3 A0A293LB34 A0A154P8H2 E0VED6 A0A151JWT9 A0A1B6IKT6 F4WK81 A0A1B6G578 A0A1B6FLE3 A0A158NIH3 A0A195BGP1 T1IUE5 A0A0N7ZBI5 J3JTU0 A0A151XJR8 A0A195C1J9 E2BUV7 A0A171A231 A0A182PA28 A0A2H8U045 M4HZ20 A0A1I9WLM6 A0A023GLZ5 T1H9Q5 G3MQD4 A0A1W4WLS5 H9N535 A0A182Q4C3 A0A165RB61 D6WFH1 A0A087UK01 A0A182U6F2 A0A182XIC8 A0A182LFG0 Q7QBX9 A0A182V4E1 F5HK76 A0A182HTB3 A0A1E1X7X7 A0A088AIM3 A0A0G3VIA2 A0A131YQE5 P20478-2 A0A1D1WAJ9 A0A023FZD1 R4WR46 A0A0A1XT45 U5EZ20 A0A2W1BMY2 A0A0A1WVL5 B4L4A8 A0A182JIW4 A0A0Q9WMW7 A0A2H1VZ23 A0A023FS93 I4DJ32 J9JML2 A0A182MEI4 L7M7P1 A0A224YQG5 B4R2U1 A0A088A2A8 V5G3M6 V9IDX6 A0A0G2QRP6 A0A084VJR5 A0A023F924 A0A1B6IB03 A0A2A3E9Y7 A0A2A4J4U6 M9V250 A0A131XHK3 A0A3R7LSZ4 A0A194QUW7 C0KJQ3 A0A1A9WZ89 A0A1I8NDX3 E2ATW3 B4JIW8 A0A182N4N2 A0A194PS35 H2BL56 K7IR04 X2JDA5 A0A182K3Y7 A0A2C9K0W2
A0A068TK10 A0A2J7RJB5 A0A0P4VPJ5 R4G5A5 Q5PRD7 A0A1Y1KMB3 V5HHJ9 A0A1Z5LAY3 A0A1J1HER3 A0A293LB34 A0A154P8H2 E0VED6 A0A151JWT9 A0A1B6IKT6 F4WK81 A0A1B6G578 A0A1B6FLE3 A0A158NIH3 A0A195BGP1 T1IUE5 A0A0N7ZBI5 J3JTU0 A0A151XJR8 A0A195C1J9 E2BUV7 A0A171A231 A0A182PA28 A0A2H8U045 M4HZ20 A0A1I9WLM6 A0A023GLZ5 T1H9Q5 G3MQD4 A0A1W4WLS5 H9N535 A0A182Q4C3 A0A165RB61 D6WFH1 A0A087UK01 A0A182U6F2 A0A182XIC8 A0A182LFG0 Q7QBX9 A0A182V4E1 F5HK76 A0A182HTB3 A0A1E1X7X7 A0A088AIM3 A0A0G3VIA2 A0A131YQE5 P20478-2 A0A1D1WAJ9 A0A023FZD1 R4WR46 A0A0A1XT45 U5EZ20 A0A2W1BMY2 A0A0A1WVL5 B4L4A8 A0A182JIW4 A0A0Q9WMW7 A0A2H1VZ23 A0A023FS93 I4DJ32 J9JML2 A0A182MEI4 L7M7P1 A0A224YQG5 B4R2U1 A0A088A2A8 V5G3M6 V9IDX6 A0A0G2QRP6 A0A084VJR5 A0A023F924 A0A1B6IB03 A0A2A3E9Y7 A0A2A4J4U6 M9V250 A0A131XHK3 A0A3R7LSZ4 A0A194QUW7 C0KJQ3 A0A1A9WZ89 A0A1I8NDX3 E2ATW3 B4JIW8 A0A182N4N2 A0A194PS35 H2BL56 K7IR04 X2JDA5 A0A182K3Y7 A0A2C9K0W2
PDB
2QC8
E-value=4.68734e-78,
Score=739
Ontologies
GO
Topology
Subcellular location
Cytoplasm
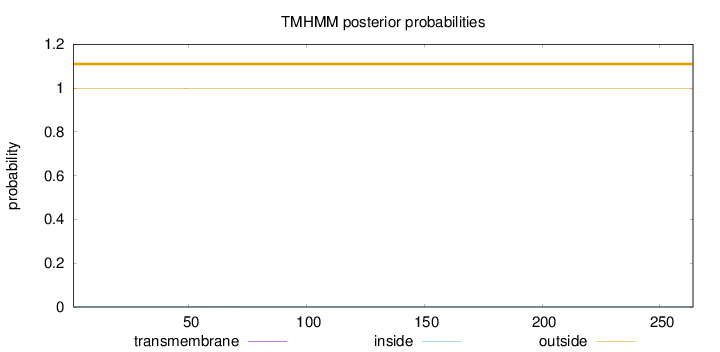
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00566
Exp number, first 60 AAs:
0.00312
Total prob of N-in:
0.00137
outside
1 - 264
Population Genetic Test Statistics
Pi
249.177616
Theta
191.098826
Tajima's D
1.014734
CLR
0.088924
CSRT
0.666666666666667
Interpretation
Uncertain
