Gene
KWMTBOMO13102
Pre Gene Modal
BGIBMGA000387
Annotation
PREDICTED:_carboxypeptidase_N_subunit_2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.512 Nuclear Reliability : 1.766
Sequence
CDS
ATGTGTAGATGCGACGCGGCCCGACGGTCACTCAACTGTTGGAGGGCCGGCCTGACTGCACTCACTGGGGACTTGACCGCGCCCGCTGATCTTCTCACCATAGATTTGGGGTCAAATAAGCTACGCACCCTACACAAGACCACATTTAAAGGAATGCGTGCGCTAACTGAACTAGATATGTTCGACAATCACGTCGAATATTTGCCGGGAGGAATATTCGAATCTCTTGTGAACTTGAAAACCCTGCGTCTACAGAGGAATTATTTAGAAGAAATTGATAGCGAAGCGTTTACGTCAACAAAAAAACTTTTCCACGTGGATTTGTCCAACAATTATTTGTATACACTACCAGAAAAGCTATTTGCCAACAATACATTTCTTGAAACCATCGATATCTCAAATAACCGTGTTGTATACGTTCCATCAGATAGTTTCTTAGGATTGTATTCTCTACAAATACTCGATTTATCGAAAAACAAAATACAAAATATCCAAAACGGAACGTTTTCGCTGAAGAACCTCCAGATCTTGAAACTGTCGGACAATAAGATAAACAACATTAGTGACAACGCCTTTGAGAACTTATTGTCCTTGGAAACTTTATTATTAGATCGGAATAAATTAAAGAATATTCCGACACAATTATTTAATAATCTGAAATGTTTGATTTTCCTAGATCTGTCCAATAATAATTTGATGAGTTTGGCTGGGGTTGAATTTGAAAGCCTGAAATTATTACGTAATTTAGATTTGAAGGAAAACGCTCTAAGCCAAATACCTGACAACACATTTTCTGACTGTTTCAACTTGGAGAAACTTGATTTATCTAAAAATAAACTACGCTTTATAAATATAACTGCTTTTTACGGTCTAGAAAATCTTACTACGCTACTTTTATCCGAAAACAAATTATATGAAGTACATTTCAAAGCGTTTTCTATGCTGAAGAATCTGACAACATTATACTTGGATGGTAATATGTTTCCTTCGCTGCCATCTCGCACCCTGGATTACATGCCGAAGCTTACGATCGTAAAATTATCTGGAAATCCTTGGCACTGCGACTGCCATGCTCTCTATATTTCAGCGTGGGTTCGTTTGAATGAAATGAAGATCTGGGATTACTCCCCAACGTGTGTATCGCCCTGGTATTTGGAAGGGCATTTCCTGAAAAAGTTAAAATTTCCAGAACTTTGCGCCGGGCAGTGGGCCAGTATGGTGAACCTTTCGCCACGCCTTCCTATACAACAGCTTCTGGCTTTAAACGTATCAGTAAATCGTAAGCCTCAAGACAGTATGGAGCAGAATCATGATTATACAACCGTCGATACCTGGCATTAA
Protein
MCRCDAARRSLNCWRAGLTALTGDLTAPADLLTIDLGSNKLRTLHKTTFKGMRALTELDMFDNHVEYLPGGIFESLVNLKTLRLQRNYLEEIDSEAFTSTKKLFHVDLSNNYLYTLPEKLFANNTFLETIDISNNRVVYVPSDSFLGLYSLQILDLSKNKIQNIQNGTFSLKNLQILKLSDNKINNISDNAFENLLSLETLLLDRNKLKNIPTQLFNNLKCLIFLDLSNNNLMSLAGVEFESLKLLRNLDLKENALSQIPDNTFSDCFNLEKLDLSKNKLRFINITAFYGLENLTTLLLSENKLYEVHFKAFSMLKNLTTLYLDGNMFPSLPSRTLDYMPKLTIVKLSGNPWHCDCHALYISAWVRLNEMKIWDYSPTCVSPWYLEGHFLKKLKFPELCAGQWASMVNLSPRLPIQQLLALNVSVNRKPQDSMEQNHDYTTVDTWH
Summary
Similarity
Belongs to the glycosyltransferase 10 family.
Uniprot
A0A2H1W9P8
H9IT08
A0A3S2LYS0
A0A212EZR9
A0A194PXX0
A0A194R6C1
+ More
D6WJ99 A0A3Q0IK40 A0A336MKV1 A0A1B6IJ18 A0A1B6BXF0 A0A1Y1MN54 A0A336LCA2 A0A2H8TF28 A0A1W4WFT5 J9JWP6 A0A1S3CVC7 A0A0A1XMP4 A0A0L0C3P3 Q29EA2 A0A3B0JBY8 A0A1W4VLU6 B4H1R0 A0A0K8VGJ9 A0A1Y1MFS4 A0A3B0JXW4 B3NG65 B4PIB8 B4QR25 A0A1B0AI55 Q9VZ84 B3M9K5 A0A1B0G8G7 B4MMX3 A0A1A9V3Q0 A0A1B0B5K1 A0A1A9Y0T6 A0A2P8Z8Y2 A0A0M4ECC9 B4IZ65 T1IBR7 A0A154PHW7 A0A0A9XNN7 B4MG03 B4KXB7 A0A0Q9VZB8 A0A0Q9XBN1 A0A1J1ICP5 A0A1A9WTK7 W8C8Y5 A0A0N8CQ19 K7J4N9 A0A0L7QKK3 A0A0P5JKY8 A0A0P6AQX8 A0A0P5C9A9 A0A0P6EVN4 A0A0P5IM68 A0A0P5G6I3 A0A0P5RQ90 A0A0P5JGR9 A0A0P5F8D9 A0A0P5EQU9 A0A310SEW6 A0A1D2NEH7 A0A146LSJ8 A0A0C9QLD7 A0A151IJ49 A0A0N8ABZ3 F4W961 W8BXB5 A0A195B4Y4 A0A088AEW7 A0A195EEN9 A0A0P6GVY6 A0A195F494 A0A0T6BFP9 A0A158P0D7 A0A151WUD5 B4HUA8 A0A0P5PRF5 E2B322 A0A0P6G9Y9 A0A3R7QWP3 E2AVJ8 A0A2S2PNT7 N6ULH3 U4U0B7 A0A2J7PFD5 A0A3L8DPQ8 A0A026WDY8 A0A164M8F0 E9G3X8 A0A0P6F9X3 A0A0P5EVF1 A0A0J7KS06 A0A0P5IMA4 A0A0P5PVP8
D6WJ99 A0A3Q0IK40 A0A336MKV1 A0A1B6IJ18 A0A1B6BXF0 A0A1Y1MN54 A0A336LCA2 A0A2H8TF28 A0A1W4WFT5 J9JWP6 A0A1S3CVC7 A0A0A1XMP4 A0A0L0C3P3 Q29EA2 A0A3B0JBY8 A0A1W4VLU6 B4H1R0 A0A0K8VGJ9 A0A1Y1MFS4 A0A3B0JXW4 B3NG65 B4PIB8 B4QR25 A0A1B0AI55 Q9VZ84 B3M9K5 A0A1B0G8G7 B4MMX3 A0A1A9V3Q0 A0A1B0B5K1 A0A1A9Y0T6 A0A2P8Z8Y2 A0A0M4ECC9 B4IZ65 T1IBR7 A0A154PHW7 A0A0A9XNN7 B4MG03 B4KXB7 A0A0Q9VZB8 A0A0Q9XBN1 A0A1J1ICP5 A0A1A9WTK7 W8C8Y5 A0A0N8CQ19 K7J4N9 A0A0L7QKK3 A0A0P5JKY8 A0A0P6AQX8 A0A0P5C9A9 A0A0P6EVN4 A0A0P5IM68 A0A0P5G6I3 A0A0P5RQ90 A0A0P5JGR9 A0A0P5F8D9 A0A0P5EQU9 A0A310SEW6 A0A1D2NEH7 A0A146LSJ8 A0A0C9QLD7 A0A151IJ49 A0A0N8ABZ3 F4W961 W8BXB5 A0A195B4Y4 A0A088AEW7 A0A195EEN9 A0A0P6GVY6 A0A195F494 A0A0T6BFP9 A0A158P0D7 A0A151WUD5 B4HUA8 A0A0P5PRF5 E2B322 A0A0P6G9Y9 A0A3R7QWP3 E2AVJ8 A0A2S2PNT7 N6ULH3 U4U0B7 A0A2J7PFD5 A0A3L8DPQ8 A0A026WDY8 A0A164M8F0 E9G3X8 A0A0P6F9X3 A0A0P5EVF1 A0A0J7KS06 A0A0P5IMA4 A0A0P5PVP8
Pubmed
19121390
22118469
26354079
18362917
19820115
28004739
+ More
25830018 26108605 15632085 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 25401762 26823975 18057021 24495485 20075255 27289101 21719571 21347285 20798317 23537049 30249741 24508170 21292972
25830018 26108605 15632085 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 25401762 26823975 18057021 24495485 20075255 27289101 21719571 21347285 20798317 23537049 30249741 24508170 21292972
EMBL
ODYU01007149
SOQ49676.1
BABH01013137
BABH01013138
BABH01013139
RSAL01000111
+ More
RVE47085.1 AGBW02011240 OWR46957.1 KQ459586 KPI97873.1 KQ460883 KPJ11411.1 KQ971343 EFA04699.1 UFQS01001567 UFQT01001567 SSX11477.1 SSX31044.1 GECU01020789 JAS86917.1 GEDC01031345 JAS05953.1 GEZM01032323 JAV84667.1 UFQS01003149 UFQT01003149 SSX15270.1 SSX34645.1 GFXV01000898 MBW12703.1 ABLF02038891 GBXI01001673 JAD12619.1 JRES01000951 KNC26861.1 CH379070 EAL30159.2 OUUW01000002 SPP77562.1 CH479203 EDW30262.1 GDHF01014336 JAI37978.1 GEZM01032322 JAV84669.1 SPP77561.1 CH954178 EDV50966.1 CM000159 EDW93460.1 CM000363 CM002912 EDX09262.1 KMY97673.1 AE014296 AY047503 AAF47941.2 AAK77235.1 CH902618 EDV39011.1 CCAG010000253 CH963847 EDW73529.2 JXJN01008760 PYGN01000143 PSN52955.1 CP012525 ALC43058.1 CH916366 EDV95587.1 ACPB03014531 ACPB03014532 KQ434902 KZC11084.1 GBHO01023166 GDHC01013266 JAG20438.1 JAQ05363.1 CH940669 EDW58264.1 KRF78205.1 CH933809 EDW17575.1 KRF78204.1 KRG05621.1 CVRI01000047 CRK98053.1 GAMC01002624 JAC03932.1 GDIP01110095 JAL93619.1 AAZX01010313 KQ414940 KOC59159.1 GDIP01158993 GDIQ01199949 GDIQ01198648 GDIQ01029273 JAJ64409.1 JAK53077.1 GDIP01031788 JAM71927.1 GDIP01173927 JAJ49475.1 GDIQ01057305 JAN37432.1 GDIQ01219398 JAK32327.1 GDIQ01267408 JAJ84316.1 GDIQ01116295 JAL35431.1 GDIQ01198647 GDIQ01093638 JAK53078.1 GDIQ01266425 JAJ85299.1 GDIQ01266424 JAJ85300.1 KQ780658 OAD51940.1 LJIJ01000067 ODN03637.1 GDHC01009229 JAQ09400.1 GBYB01004339 JAG74106.1 KQ977412 KYN02904.1 GDIP01162393 JAJ61009.1 GL888002 EGI69243.1 GAMC01002623 JAC03933.1 KQ976598 KYM79563.1 KQ979039 KYN23272.1 GDIQ01027701 JAN67036.1 KQ981855 KYN34904.1 LJIG01000762 KRT86178.1 ADTU01000983 ADTU01000984 KQ982736 KYQ51463.1 CH480817 EDW50529.1 GDIP01240129 GDIQ01124425 JAI83272.1 JAL27301.1 GL445255 EFN89908.1 GDIQ01039118 JAN55619.1 QCYY01001043 ROT80960.1 GL443122 EFN62549.1 GGMR01018490 MBY31109.1 APGK01028341 KB740686 ENN79552.1 KB631930 ERL87309.1 NEVH01025672 PNF15037.1 QOIP01000005 RLU22276.1 KK107293 EZA53244.1 LRGB01003056 KZS04830.1 GL732531 EFX85909.1 GDIQ01050871 JAN43866.1 GDIQ01267406 JAJ84318.1 LBMM01003761 KMQ93167.1 GDIQ01236130 JAK15595.1 GDIQ01133128 JAL18598.1
RVE47085.1 AGBW02011240 OWR46957.1 KQ459586 KPI97873.1 KQ460883 KPJ11411.1 KQ971343 EFA04699.1 UFQS01001567 UFQT01001567 SSX11477.1 SSX31044.1 GECU01020789 JAS86917.1 GEDC01031345 JAS05953.1 GEZM01032323 JAV84667.1 UFQS01003149 UFQT01003149 SSX15270.1 SSX34645.1 GFXV01000898 MBW12703.1 ABLF02038891 GBXI01001673 JAD12619.1 JRES01000951 KNC26861.1 CH379070 EAL30159.2 OUUW01000002 SPP77562.1 CH479203 EDW30262.1 GDHF01014336 JAI37978.1 GEZM01032322 JAV84669.1 SPP77561.1 CH954178 EDV50966.1 CM000159 EDW93460.1 CM000363 CM002912 EDX09262.1 KMY97673.1 AE014296 AY047503 AAF47941.2 AAK77235.1 CH902618 EDV39011.1 CCAG010000253 CH963847 EDW73529.2 JXJN01008760 PYGN01000143 PSN52955.1 CP012525 ALC43058.1 CH916366 EDV95587.1 ACPB03014531 ACPB03014532 KQ434902 KZC11084.1 GBHO01023166 GDHC01013266 JAG20438.1 JAQ05363.1 CH940669 EDW58264.1 KRF78205.1 CH933809 EDW17575.1 KRF78204.1 KRG05621.1 CVRI01000047 CRK98053.1 GAMC01002624 JAC03932.1 GDIP01110095 JAL93619.1 AAZX01010313 KQ414940 KOC59159.1 GDIP01158993 GDIQ01199949 GDIQ01198648 GDIQ01029273 JAJ64409.1 JAK53077.1 GDIP01031788 JAM71927.1 GDIP01173927 JAJ49475.1 GDIQ01057305 JAN37432.1 GDIQ01219398 JAK32327.1 GDIQ01267408 JAJ84316.1 GDIQ01116295 JAL35431.1 GDIQ01198647 GDIQ01093638 JAK53078.1 GDIQ01266425 JAJ85299.1 GDIQ01266424 JAJ85300.1 KQ780658 OAD51940.1 LJIJ01000067 ODN03637.1 GDHC01009229 JAQ09400.1 GBYB01004339 JAG74106.1 KQ977412 KYN02904.1 GDIP01162393 JAJ61009.1 GL888002 EGI69243.1 GAMC01002623 JAC03933.1 KQ976598 KYM79563.1 KQ979039 KYN23272.1 GDIQ01027701 JAN67036.1 KQ981855 KYN34904.1 LJIG01000762 KRT86178.1 ADTU01000983 ADTU01000984 KQ982736 KYQ51463.1 CH480817 EDW50529.1 GDIP01240129 GDIQ01124425 JAI83272.1 JAL27301.1 GL445255 EFN89908.1 GDIQ01039118 JAN55619.1 QCYY01001043 ROT80960.1 GL443122 EFN62549.1 GGMR01018490 MBY31109.1 APGK01028341 KB740686 ENN79552.1 KB631930 ERL87309.1 NEVH01025672 PNF15037.1 QOIP01000005 RLU22276.1 KK107293 EZA53244.1 LRGB01003056 KZS04830.1 GL732531 EFX85909.1 GDIQ01050871 JAN43866.1 GDIQ01267406 JAJ84318.1 LBMM01003761 KMQ93167.1 GDIQ01236130 JAK15595.1 GDIQ01133128 JAL18598.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000007266
+ More
UP000079169 UP000192223 UP000007819 UP000037069 UP000001819 UP000268350 UP000192221 UP000008744 UP000008711 UP000002282 UP000000304 UP000092445 UP000000803 UP000007801 UP000092444 UP000007798 UP000078200 UP000092460 UP000092443 UP000245037 UP000092553 UP000001070 UP000015103 UP000076502 UP000008792 UP000009192 UP000183832 UP000091820 UP000002358 UP000053825 UP000094527 UP000078542 UP000007755 UP000078540 UP000005203 UP000078492 UP000078541 UP000005205 UP000075809 UP000001292 UP000008237 UP000283509 UP000000311 UP000019118 UP000030742 UP000235965 UP000279307 UP000053097 UP000076858 UP000000305 UP000036403
UP000079169 UP000192223 UP000007819 UP000037069 UP000001819 UP000268350 UP000192221 UP000008744 UP000008711 UP000002282 UP000000304 UP000092445 UP000000803 UP000007801 UP000092444 UP000007798 UP000078200 UP000092460 UP000092443 UP000245037 UP000092553 UP000001070 UP000015103 UP000076502 UP000008792 UP000009192 UP000183832 UP000091820 UP000002358 UP000053825 UP000094527 UP000078542 UP000007755 UP000078540 UP000005203 UP000078492 UP000078541 UP000005205 UP000075809 UP000001292 UP000008237 UP000283509 UP000000311 UP000019118 UP000030742 UP000235965 UP000279307 UP000053097 UP000076858 UP000000305 UP000036403
Pfam
Interpro
IPR032675
LRR_dom_sf
+ More
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR020894 Cadherin_CS
IPR000225 Armadillo
IPR002126 Cadherin-like_dom
IPR026906 LRR_5
IPR000483 Cys-rich_flank_reg_C
IPR025875 Leu-rich_rpt_4
IPR006170 PBP/GOBP
IPR036728 PBP_GOBP_sf
IPR031283 AMIGO
IPR038577 GT10-like_sf
IPR001503 Glyco_trans_10
IPR031481 Glyco_tran_10_N
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR020894 Cadherin_CS
IPR000225 Armadillo
IPR002126 Cadherin-like_dom
IPR026906 LRR_5
IPR000483 Cys-rich_flank_reg_C
IPR025875 Leu-rich_rpt_4
IPR006170 PBP/GOBP
IPR036728 PBP_GOBP_sf
IPR031283 AMIGO
IPR038577 GT10-like_sf
IPR001503 Glyco_trans_10
IPR031481 Glyco_tran_10_N
SUPFAM
SSF47565
SSF47565
Gene 3D
ProteinModelPortal
A0A2H1W9P8
H9IT08
A0A3S2LYS0
A0A212EZR9
A0A194PXX0
A0A194R6C1
+ More
D6WJ99 A0A3Q0IK40 A0A336MKV1 A0A1B6IJ18 A0A1B6BXF0 A0A1Y1MN54 A0A336LCA2 A0A2H8TF28 A0A1W4WFT5 J9JWP6 A0A1S3CVC7 A0A0A1XMP4 A0A0L0C3P3 Q29EA2 A0A3B0JBY8 A0A1W4VLU6 B4H1R0 A0A0K8VGJ9 A0A1Y1MFS4 A0A3B0JXW4 B3NG65 B4PIB8 B4QR25 A0A1B0AI55 Q9VZ84 B3M9K5 A0A1B0G8G7 B4MMX3 A0A1A9V3Q0 A0A1B0B5K1 A0A1A9Y0T6 A0A2P8Z8Y2 A0A0M4ECC9 B4IZ65 T1IBR7 A0A154PHW7 A0A0A9XNN7 B4MG03 B4KXB7 A0A0Q9VZB8 A0A0Q9XBN1 A0A1J1ICP5 A0A1A9WTK7 W8C8Y5 A0A0N8CQ19 K7J4N9 A0A0L7QKK3 A0A0P5JKY8 A0A0P6AQX8 A0A0P5C9A9 A0A0P6EVN4 A0A0P5IM68 A0A0P5G6I3 A0A0P5RQ90 A0A0P5JGR9 A0A0P5F8D9 A0A0P5EQU9 A0A310SEW6 A0A1D2NEH7 A0A146LSJ8 A0A0C9QLD7 A0A151IJ49 A0A0N8ABZ3 F4W961 W8BXB5 A0A195B4Y4 A0A088AEW7 A0A195EEN9 A0A0P6GVY6 A0A195F494 A0A0T6BFP9 A0A158P0D7 A0A151WUD5 B4HUA8 A0A0P5PRF5 E2B322 A0A0P6G9Y9 A0A3R7QWP3 E2AVJ8 A0A2S2PNT7 N6ULH3 U4U0B7 A0A2J7PFD5 A0A3L8DPQ8 A0A026WDY8 A0A164M8F0 E9G3X8 A0A0P6F9X3 A0A0P5EVF1 A0A0J7KS06 A0A0P5IMA4 A0A0P5PVP8
D6WJ99 A0A3Q0IK40 A0A336MKV1 A0A1B6IJ18 A0A1B6BXF0 A0A1Y1MN54 A0A336LCA2 A0A2H8TF28 A0A1W4WFT5 J9JWP6 A0A1S3CVC7 A0A0A1XMP4 A0A0L0C3P3 Q29EA2 A0A3B0JBY8 A0A1W4VLU6 B4H1R0 A0A0K8VGJ9 A0A1Y1MFS4 A0A3B0JXW4 B3NG65 B4PIB8 B4QR25 A0A1B0AI55 Q9VZ84 B3M9K5 A0A1B0G8G7 B4MMX3 A0A1A9V3Q0 A0A1B0B5K1 A0A1A9Y0T6 A0A2P8Z8Y2 A0A0M4ECC9 B4IZ65 T1IBR7 A0A154PHW7 A0A0A9XNN7 B4MG03 B4KXB7 A0A0Q9VZB8 A0A0Q9XBN1 A0A1J1ICP5 A0A1A9WTK7 W8C8Y5 A0A0N8CQ19 K7J4N9 A0A0L7QKK3 A0A0P5JKY8 A0A0P6AQX8 A0A0P5C9A9 A0A0P6EVN4 A0A0P5IM68 A0A0P5G6I3 A0A0P5RQ90 A0A0P5JGR9 A0A0P5F8D9 A0A0P5EQU9 A0A310SEW6 A0A1D2NEH7 A0A146LSJ8 A0A0C9QLD7 A0A151IJ49 A0A0N8ABZ3 F4W961 W8BXB5 A0A195B4Y4 A0A088AEW7 A0A195EEN9 A0A0P6GVY6 A0A195F494 A0A0T6BFP9 A0A158P0D7 A0A151WUD5 B4HUA8 A0A0P5PRF5 E2B322 A0A0P6G9Y9 A0A3R7QWP3 E2AVJ8 A0A2S2PNT7 N6ULH3 U4U0B7 A0A2J7PFD5 A0A3L8DPQ8 A0A026WDY8 A0A164M8F0 E9G3X8 A0A0P6F9X3 A0A0P5EVF1 A0A0J7KS06 A0A0P5IMA4 A0A0P5PVP8
PDB
5A5C
E-value=1.83458e-29,
Score=323
Ontologies
GO
PANTHER
Topology
Subcellular location
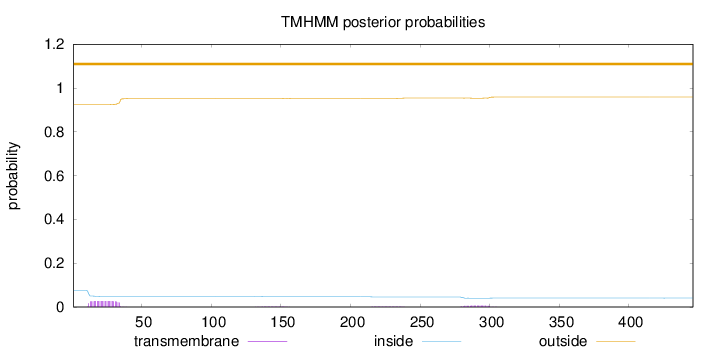
Length:
446
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.78067
Exp number, first 60 AAs:
0.59677
Total prob of N-in:
0.07562
outside
1 - 446
Population Genetic Test Statistics
Pi
282.933698
Theta
190.313512
Tajima's D
1.605651
CLR
0.362206
CSRT
0.810309484525774
Interpretation
Uncertain
