Gene
KWMTBOMO13094 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000390
Annotation
PREDICTED:_phosphoribosylformylglycinamidine_synthase_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.226
Sequence
CDS
ATGCCACGAAAAACCTTCGATTTACAAACTGAAAAAAGGACCAAGTTACCTTTGTCATTTCCTAAAGACATAACGGTGAAGAGTGCACTCGATCGCGTTCTGAGGCTTGTCAATGTTGCCAGCAAAAGATACCTAACCAACAAGGTCGACCGTTGTGTCACTGGTCTCGTAGCCCAACAGCAATGTGTGGGCCCACTTCACACTCCGCTGGCGGATTGCGCTATTATAGCACTTTCTTACTACGACCTCGTCGGCTCGGCCACTTCTATCGGTACACAGAACATCAAGGGACTACTGGACCCGGCAGCCGGAGCGAGATTGTCATTAGGGGAGGCTCTAACTAATTTGGTGTTCGCCGGCATATCGGAGCTGGAGGACGTGAAGTGTTCAGGGAACTGGATGTGGGGCGGCAAGACCGGCGTGGAGGGCGGCGCGCTGACCGCGGCGTGCGGCGCGCTGTGCGGCGCCATGCGCGCGCTCGGCGTCGCCGTCGACGGCGGGAAGGACTCGCTCTCCATGTGCGCACACGTCGCCGGCGAGAAAGTTAAGTCTCCGGGAACTTTGGTGGTATCGACATACGCCCCTTGTCCAGATATTACCGTTAAAATTGAACCAGCACTCAAAGAAGAAAACTCCGCTCTGGTTCACGCACCTGTTTCACCAGGAAAATACAGACTTGGTGGTTCAGCATTGGCTCAATGTTACAAGCAGCTAGGCGATAATCCTCCCGATTTAGACGATCCCATCATATTGAAAAGTCTATTCAAGATTACACAGAAATTACTAAAAGAAAAAAAATTGATATCCGGTCATGATATCAGCGAAGGAGGCTTCATAACAACGGTACTTGAGATGGGTATCGGTGGTGTTCGCGGTTTGAATCTACACGTCAACGTTGCCGCCAATGTCTCGCCCATCGAAGCGCTGTTCAATGAGGAACTTGGTATCGTTCTGGAAGTTTCCAAAAACGACTTGCCCTATGTTCTAAGCGAATACAACAACAATGGCGTCGGTGCACAGCACATTGGTTACACTGGGAAATACGGCATGGATTCTCAGGTTGTGATAAATTTGAATGGCGTTAATGTTTTGGATACTGTTCTCATTGATGTGTTCAGAATGTGGGAGGAAACTAGTTATCGTTTGGAATGCTTGCAAGCTAACACTGATTGTGTCCTCCAAGAATGGAAAGTTCTTGAGAAACGTAAAGGAGCCACATACAACGTTACTTTTGATCCTACGGCTGCAGTTATTAAAACGAAATCCATTAAGGTTGCAGTACTTCGTGAAGAAGGTATCAATGGAGACAGGGAAATGATCGCATCTCTAATGCTAGCCAACTTTGATGTGTTTGATGTAACAATGAGTGACTTACAAAACAAGAAGATTACTCTGGATGCGTTCCAAGGTGTTGTTTTCCCGGGAGGTTTCAGTTACGCAGACACTCTGGGTTCTGCTAAAGGCTGGGCTGCTGGAATATTATTCTCAGAATCTCTAAGCTCGCAATTTGCTCACTTTAAAGATAGAAAAGACACTTTCTCTTTGGGAGTTTGTAATGGGTGTCAGCTAATGGCTATGCTAGGATGGGTGGATCCCGATAGCAATTCCAAAGGTTCAAACAAGGCCCAGATATTCCTAGACCATAATAGCTCGGAAAGATTTGAATGCCGATGGAGTGCCGTGAAGATACACGAGAACGAGAACAAAGAAGACGTGTGGTTCCGCGGGATGGGCGGCAGCGTGCTCGGCGTGTGGGTCGCGCACGGCGAAGGACGATTCACGGTCGCCGACGGTTCCCTACTCGACAAACTGTACAAGAACGGACAAGTCGCTGTACAGTATGTCGACGACGACGGAAAGCCCACCGAGGTGTACCCGATGAATCCCAATGGAAGTCCAGATGGGCTGGCTGGTGTCCGTTCACGTGACGGCCGTCACATAGCTATGATGCCACATCCAGAGCGTTGTGTTTTACGTTGGCAATGTCCGGCTCCTTGCCCCACGGTAACTAATCCGTCTGTACGCAACAGCCAAGCCTCGCCGTGGTTGCGGCTCTTCCAAAATGCCTACTCGTGGGCTATTAATCATTAA
Protein
MPRKTFDLQTEKRTKLPLSFPKDITVKSALDRVLRLVNVASKRYLTNKVDRCVTGLVAQQQCVGPLHTPLADCAIIALSYYDLVGSATSIGTQNIKGLLDPAAGARLSLGEALTNLVFAGISELEDVKCSGNWMWGGKTGVEGGALTAACGALCGAMRALGVAVDGGKDSLSMCAHVAGEKVKSPGTLVVSTYAPCPDITVKIEPALKEENSALVHAPVSPGKYRLGGSALAQCYKQLGDNPPDLDDPIILKSLFKITQKLLKEKKLISGHDISEGGFITTVLEMGIGGVRGLNLHVNVAANVSPIEALFNEELGIVLEVSKNDLPYVLSEYNNNGVGAQHIGYTGKYGMDSQVVINLNGVNVLDTVLIDVFRMWEETSYRLECLQANTDCVLQEWKVLEKRKGATYNVTFDPTAAVIKTKSIKVAVLREEGINGDREMIASLMLANFDVFDVTMSDLQNKKITLDAFQGVVFPGGFSYADTLGSAKGWAAGILFSESLSSQFAHFKDRKDTFSLGVCNGCQLMAMLGWVDPDSNSKGSNKAQIFLDHNSSERFECRWSAVKIHENENKEDVWFRGMGGSVLGVWVAHGEGRFTVADGSLLDKLYKNGQVAVQYVDDDGKPTEVYPMNPNGSPDGLAGVRSRDGRHIAMMPHPERCVLRWQCPAPCPTVTNPSVRNSQASPWLRLFQNAYSWAINH
Summary
Uniprot
H9IT11
A0A2H1VMD1
A0A2W1BTP0
A0A194PX17
A0A212EZS4
A0A194R0U5
+ More
K7J2R4 A0A1Z5L8M9 E0V9E3 A0A226E6D9 A0A087UDD0 A0A232F240 A0A067R9T8 A0A2R5L6G5 A0A310SK93 A0A0C9QLK2 A0A195FTM4 A0A0C9QQD9 A0A293N034 A0A151IIZ1 A0A0L7QQW1 A0A1V9Y1C1 F4WA86 A0A2A3E8W2 A0A1D2N9H3 C3XX99 A0A151JP83 A0A088A349 A0A1L8ERG1 V5HDX7 A0A195B5R5 A0A2J7RH08 A0A131Y2I7 A0A0N0BDG7 A0A2J7RGZ3 A0A158NQD6 K9J888 A0A2J7RGZ5 A0A3Q1AM71 A0A0J7KQ86 B5DEE8 A0A151WGG9 A0A2P6KZ25 A0A0L0BT40 L7N3J6 I3J3D9 A0A1L8DXU7 A0A1I8NMG1 A0A1Y1MYP2 A0A146NQ78 A0A146RXX8 A0A026WRS5 A0A3Q3C8V8 H9G7M9 A0A1W4X6B4 A0A3L8DNY1 A0A146PF27 A0A146PMU9 A0A3Q1G0G1 A0A3Q1FVB7 A0A131Z1A9 A0A0P4VVS8 A0A1E1X997 A0A3Q2Q3V2 A0A3Q4I9L2 A0A0T6B804 A0A336M968 A0A3P9CVQ5 A0A3Q2NMR5 A0A3Q2CY86 A0A3Q1FV88 A0A3P9N2J3 A0A224Z4A2 A0A2C9JR22 A0A146SCI4 A0A336MBE0 A0A3Q1C0E8 A0A3B3TIN3 N6TFV9 A0A3B5LQU2 A0A3Q1J1K8 A0A315VNE6 W4VRR3 A0A3B3WI83 A0A3P8SVX2 A0A210QX06 A0A3Q3SJQ7 A0A3P8PIL4 A0A3Q3MLE8 A0A087Y6E0 A0A3Q3AGK5 T1DLT2 A0A2L2Y2M0 Q17A67 A0A1A7YI05 R7TFX7 A0A1A7XQ43 X1ZKR9 A0A3B5AQV9 A0A146M9G1
K7J2R4 A0A1Z5L8M9 E0V9E3 A0A226E6D9 A0A087UDD0 A0A232F240 A0A067R9T8 A0A2R5L6G5 A0A310SK93 A0A0C9QLK2 A0A195FTM4 A0A0C9QQD9 A0A293N034 A0A151IIZ1 A0A0L7QQW1 A0A1V9Y1C1 F4WA86 A0A2A3E8W2 A0A1D2N9H3 C3XX99 A0A151JP83 A0A088A349 A0A1L8ERG1 V5HDX7 A0A195B5R5 A0A2J7RH08 A0A131Y2I7 A0A0N0BDG7 A0A2J7RGZ3 A0A158NQD6 K9J888 A0A2J7RGZ5 A0A3Q1AM71 A0A0J7KQ86 B5DEE8 A0A151WGG9 A0A2P6KZ25 A0A0L0BT40 L7N3J6 I3J3D9 A0A1L8DXU7 A0A1I8NMG1 A0A1Y1MYP2 A0A146NQ78 A0A146RXX8 A0A026WRS5 A0A3Q3C8V8 H9G7M9 A0A1W4X6B4 A0A3L8DNY1 A0A146PF27 A0A146PMU9 A0A3Q1G0G1 A0A3Q1FVB7 A0A131Z1A9 A0A0P4VVS8 A0A1E1X997 A0A3Q2Q3V2 A0A3Q4I9L2 A0A0T6B804 A0A336M968 A0A3P9CVQ5 A0A3Q2NMR5 A0A3Q2CY86 A0A3Q1FV88 A0A3P9N2J3 A0A224Z4A2 A0A2C9JR22 A0A146SCI4 A0A336MBE0 A0A3Q1C0E8 A0A3B3TIN3 N6TFV9 A0A3B5LQU2 A0A3Q1J1K8 A0A315VNE6 W4VRR3 A0A3B3WI83 A0A3P8SVX2 A0A210QX06 A0A3Q3SJQ7 A0A3P8PIL4 A0A3Q3MLE8 A0A087Y6E0 A0A3Q3AGK5 T1DLT2 A0A2L2Y2M0 Q17A67 A0A1A7YI05 R7TFX7 A0A1A7XQ43 X1ZKR9 A0A3B5AQV9 A0A146M9G1
Pubmed
19121390
28756777
26354079
22118469
20075255
28528879
+ More
20566863 28648823 24845553 28327890 21719571 27289101 18563158 27762356 25765539 21347285 20431018 26108605 25186727 28004739 24508170 21881562 30249741 26830274 27129103 28503490 28797301 15562597 23537049 29703783 28812685 23758969 26561354 17510324 23254933 26823975
20566863 28648823 24845553 28327890 21719571 27289101 18563158 27762356 25765539 21347285 20431018 26108605 25186727 28004739 24508170 21881562 30249741 26830274 27129103 28503490 28797301 15562597 23537049 29703783 28812685 23758969 26561354 17510324 23254933 26823975
EMBL
BABH01013153
BABH01013154
ODYU01003346
SOQ41989.1
KZ150009
PZC75103.1
+ More
KQ459586 KPI97867.1 AGBW02011240 OWR46951.1 KQ460883 KPJ11418.1 GFJQ02003241 JAW03729.1 DS234992 EEB09999.1 LNIX01000006 OXA53192.1 KK119317 KFM75369.1 NNAY01001258 OXU24590.1 KK852794 KDR16414.1 GGLE01000970 MBY05096.1 KQ765605 OAD53852.1 GBYB01001517 JAG71284.1 KQ981272 KYN43813.1 GBYB01002847 JAG72614.1 GFWV01020212 MAA44940.1 KQ977412 KYN02867.1 KQ414786 KOC60876.1 MNPL01000952 OQR79567.1 GL888045 EGI68877.1 KZ288322 PBC28158.1 LJIJ01000134 ODN01910.1 GG666471 EEN67359.1 KQ978756 KYN28592.1 CM004483 OCT61927.1 GANP01008789 JAB75679.1 KQ976595 KYM79602.1 NEVH01003749 PNF40112.1 GEFM01003104 JAP72692.1 KQ435878 KOX70064.1 PNF40114.1 ADTU01023073 ADTU01023074 ADTU01023075 ADTU01023076 ADTU01023077 ADTU01023078 ADTU01023079 AAMC01096052 PNF40115.1 LBMM01004407 KMQ92446.1 BC168641 AAI68641.1 KQ983176 KYQ46907.1 MWRG01003428 PRD31579.1 JRES01001393 KNC23191.1 AERX01004326 GFDF01002811 JAV11273.1 GEZM01017306 GEZM01017305 JAV90783.1 GCES01152740 JAQ33582.1 GCES01113131 JAQ73191.1 KK107128 EZA58391.1 AAWZ02028504 AAWZ02028505 QOIP01000006 RLU21996.1 GCES01143987 JAQ42335.1 GCES01141215 JAQ45107.1 GEDV01003975 JAP84582.1 GDKW01000428 JAI56167.1 GFAC01003355 JAT95833.1 LJIG01009308 KRT83339.1 UFQT01000742 SSX26872.1 GFPF01010615 MAA21761.1 GCES01108011 JAQ78311.1 UFQT01000651 SSX26123.1 APGK01039519 APGK01039520 KB740971 ENN76633.1 NHOQ01001926 PWA20761.1 GANO01000762 JAB59109.1 NEDP02001454 OWF53251.1 AYCK01004012 GAAZ01001882 JAA96061.1 IAAA01001579 LAA02401.1 CH477339 EAT43159.1 HADX01007980 SBP30212.1 KB310029 ELT92693.1 HADW01018535 SBP19935.1 AMQN01013186 AMQN01013187 GDHC01003429 JAQ15200.1
KQ459586 KPI97867.1 AGBW02011240 OWR46951.1 KQ460883 KPJ11418.1 GFJQ02003241 JAW03729.1 DS234992 EEB09999.1 LNIX01000006 OXA53192.1 KK119317 KFM75369.1 NNAY01001258 OXU24590.1 KK852794 KDR16414.1 GGLE01000970 MBY05096.1 KQ765605 OAD53852.1 GBYB01001517 JAG71284.1 KQ981272 KYN43813.1 GBYB01002847 JAG72614.1 GFWV01020212 MAA44940.1 KQ977412 KYN02867.1 KQ414786 KOC60876.1 MNPL01000952 OQR79567.1 GL888045 EGI68877.1 KZ288322 PBC28158.1 LJIJ01000134 ODN01910.1 GG666471 EEN67359.1 KQ978756 KYN28592.1 CM004483 OCT61927.1 GANP01008789 JAB75679.1 KQ976595 KYM79602.1 NEVH01003749 PNF40112.1 GEFM01003104 JAP72692.1 KQ435878 KOX70064.1 PNF40114.1 ADTU01023073 ADTU01023074 ADTU01023075 ADTU01023076 ADTU01023077 ADTU01023078 ADTU01023079 AAMC01096052 PNF40115.1 LBMM01004407 KMQ92446.1 BC168641 AAI68641.1 KQ983176 KYQ46907.1 MWRG01003428 PRD31579.1 JRES01001393 KNC23191.1 AERX01004326 GFDF01002811 JAV11273.1 GEZM01017306 GEZM01017305 JAV90783.1 GCES01152740 JAQ33582.1 GCES01113131 JAQ73191.1 KK107128 EZA58391.1 AAWZ02028504 AAWZ02028505 QOIP01000006 RLU21996.1 GCES01143987 JAQ42335.1 GCES01141215 JAQ45107.1 GEDV01003975 JAP84582.1 GDKW01000428 JAI56167.1 GFAC01003355 JAT95833.1 LJIG01009308 KRT83339.1 UFQT01000742 SSX26872.1 GFPF01010615 MAA21761.1 GCES01108011 JAQ78311.1 UFQT01000651 SSX26123.1 APGK01039519 APGK01039520 KB740971 ENN76633.1 NHOQ01001926 PWA20761.1 GANO01000762 JAB59109.1 NEDP02001454 OWF53251.1 AYCK01004012 GAAZ01001882 JAA96061.1 IAAA01001579 LAA02401.1 CH477339 EAT43159.1 HADX01007980 SBP30212.1 KB310029 ELT92693.1 HADW01018535 SBP19935.1 AMQN01013186 AMQN01013187 GDHC01003429 JAQ15200.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000002358
UP000009046
+ More
UP000198287 UP000054359 UP000215335 UP000027135 UP000078541 UP000078542 UP000053825 UP000192247 UP000007755 UP000242457 UP000094527 UP000001554 UP000078492 UP000005203 UP000186698 UP000078540 UP000235965 UP000053105 UP000005205 UP000008143 UP000257160 UP000036403 UP000075809 UP000037069 UP000005207 UP000095300 UP000053097 UP000264840 UP000001646 UP000192223 UP000279307 UP000257200 UP000265000 UP000261580 UP000265160 UP000265020 UP000242638 UP000076420 UP000261500 UP000019118 UP000261380 UP000265040 UP000261480 UP000265080 UP000242188 UP000261640 UP000265100 UP000028760 UP000264800 UP000008820 UP000014760 UP000261400
UP000198287 UP000054359 UP000215335 UP000027135 UP000078541 UP000078542 UP000053825 UP000192247 UP000007755 UP000242457 UP000094527 UP000001554 UP000078492 UP000005203 UP000186698 UP000078540 UP000235965 UP000053105 UP000005205 UP000008143 UP000257160 UP000036403 UP000075809 UP000037069 UP000005207 UP000095300 UP000053097 UP000264840 UP000001646 UP000192223 UP000279307 UP000257200 UP000265000 UP000261580 UP000265160 UP000265020 UP000242638 UP000076420 UP000261500 UP000019118 UP000261380 UP000265040 UP000261480 UP000265080 UP000242188 UP000261640 UP000265100 UP000028760 UP000264800 UP000008820 UP000014760 UP000261400
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IT11
A0A2H1VMD1
A0A2W1BTP0
A0A194PX17
A0A212EZS4
A0A194R0U5
+ More
K7J2R4 A0A1Z5L8M9 E0V9E3 A0A226E6D9 A0A087UDD0 A0A232F240 A0A067R9T8 A0A2R5L6G5 A0A310SK93 A0A0C9QLK2 A0A195FTM4 A0A0C9QQD9 A0A293N034 A0A151IIZ1 A0A0L7QQW1 A0A1V9Y1C1 F4WA86 A0A2A3E8W2 A0A1D2N9H3 C3XX99 A0A151JP83 A0A088A349 A0A1L8ERG1 V5HDX7 A0A195B5R5 A0A2J7RH08 A0A131Y2I7 A0A0N0BDG7 A0A2J7RGZ3 A0A158NQD6 K9J888 A0A2J7RGZ5 A0A3Q1AM71 A0A0J7KQ86 B5DEE8 A0A151WGG9 A0A2P6KZ25 A0A0L0BT40 L7N3J6 I3J3D9 A0A1L8DXU7 A0A1I8NMG1 A0A1Y1MYP2 A0A146NQ78 A0A146RXX8 A0A026WRS5 A0A3Q3C8V8 H9G7M9 A0A1W4X6B4 A0A3L8DNY1 A0A146PF27 A0A146PMU9 A0A3Q1G0G1 A0A3Q1FVB7 A0A131Z1A9 A0A0P4VVS8 A0A1E1X997 A0A3Q2Q3V2 A0A3Q4I9L2 A0A0T6B804 A0A336M968 A0A3P9CVQ5 A0A3Q2NMR5 A0A3Q2CY86 A0A3Q1FV88 A0A3P9N2J3 A0A224Z4A2 A0A2C9JR22 A0A146SCI4 A0A336MBE0 A0A3Q1C0E8 A0A3B3TIN3 N6TFV9 A0A3B5LQU2 A0A3Q1J1K8 A0A315VNE6 W4VRR3 A0A3B3WI83 A0A3P8SVX2 A0A210QX06 A0A3Q3SJQ7 A0A3P8PIL4 A0A3Q3MLE8 A0A087Y6E0 A0A3Q3AGK5 T1DLT2 A0A2L2Y2M0 Q17A67 A0A1A7YI05 R7TFX7 A0A1A7XQ43 X1ZKR9 A0A3B5AQV9 A0A146M9G1
K7J2R4 A0A1Z5L8M9 E0V9E3 A0A226E6D9 A0A087UDD0 A0A232F240 A0A067R9T8 A0A2R5L6G5 A0A310SK93 A0A0C9QLK2 A0A195FTM4 A0A0C9QQD9 A0A293N034 A0A151IIZ1 A0A0L7QQW1 A0A1V9Y1C1 F4WA86 A0A2A3E8W2 A0A1D2N9H3 C3XX99 A0A151JP83 A0A088A349 A0A1L8ERG1 V5HDX7 A0A195B5R5 A0A2J7RH08 A0A131Y2I7 A0A0N0BDG7 A0A2J7RGZ3 A0A158NQD6 K9J888 A0A2J7RGZ5 A0A3Q1AM71 A0A0J7KQ86 B5DEE8 A0A151WGG9 A0A2P6KZ25 A0A0L0BT40 L7N3J6 I3J3D9 A0A1L8DXU7 A0A1I8NMG1 A0A1Y1MYP2 A0A146NQ78 A0A146RXX8 A0A026WRS5 A0A3Q3C8V8 H9G7M9 A0A1W4X6B4 A0A3L8DNY1 A0A146PF27 A0A146PMU9 A0A3Q1G0G1 A0A3Q1FVB7 A0A131Z1A9 A0A0P4VVS8 A0A1E1X997 A0A3Q2Q3V2 A0A3Q4I9L2 A0A0T6B804 A0A336M968 A0A3P9CVQ5 A0A3Q2NMR5 A0A3Q2CY86 A0A3Q1FV88 A0A3P9N2J3 A0A224Z4A2 A0A2C9JR22 A0A146SCI4 A0A336MBE0 A0A3Q1C0E8 A0A3B3TIN3 N6TFV9 A0A3B5LQU2 A0A3Q1J1K8 A0A315VNE6 W4VRR3 A0A3B3WI83 A0A3P8SVX2 A0A210QX06 A0A3Q3SJQ7 A0A3P8PIL4 A0A3Q3MLE8 A0A087Y6E0 A0A3Q3AGK5 T1DLT2 A0A2L2Y2M0 Q17A67 A0A1A7YI05 R7TFX7 A0A1A7XQ43 X1ZKR9 A0A3B5AQV9 A0A146M9G1
PDB
4R7G
E-value=4.85507e-112,
Score=1037
Ontologies
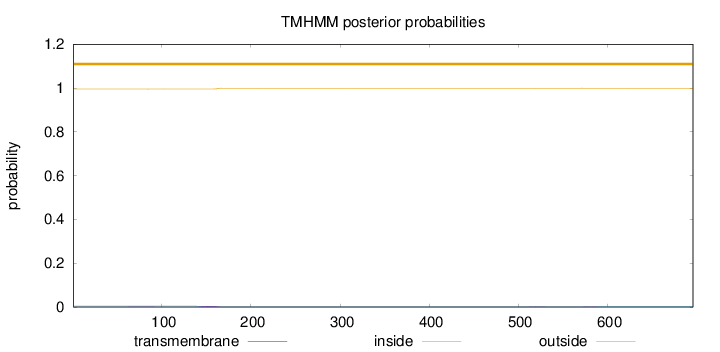
Topology
Length:
696
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1082
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00485
outside
1 - 696
Population Genetic Test Statistics
Pi
280.398537
Theta
182.268034
Tajima's D
2.040444
CLR
0
CSRT
0.891255437228139
Interpretation
Uncertain
