Gene
KWMTBOMO13087
Pre Gene Modal
BGIBMGA000308
Annotation
PREDICTED:_synapsin_[Bombyx_mori]
Full name
Synapsin
Location in the cell
Nuclear Reliability : 3.411
Sequence
CDS
ATGAAGATTAGTTCTTTATATTTAATAGATAACTTTGGTAGGATTAAAAGTTATAGGTGTAAAATCGTGAGTGGCTACATAGTGTGTGTCGGTTTTGTCCCTCCTGTACGTGTCCCTGTTTGGTGGCGTGGCGGTGGGGCGCGTAGTTCAGTGGAGGCATTGCGCAGATTCAGCTCCGGTGATCTGCAGTCGGAGTGCGATGAGGGCGAGGTGGACCCTTACACGGGCCGCGCGGCCACCACTAGTCAACCCAAGCCGCAAGCTCAAAAGCCCACGTCGCAGGCGACTGGAGGCAGCTTGGGAGGCTCGTCCGGACCTCCGCCAGCGCCGTCTGCTGCAGCAGCTGGTGGGGACCTCTCTCTTAATTTGCGAGGCGGCTCTAGGGGTACATCAGCGCCATCTTCCCCCGCCAAATCTAGAGAGTCCTTGTTGCAACGGGTACAATCTCTCACAGGCGCCGCTCGTGATCAAGGCGCTAATTTACTCGGAAGTGTAACATCGGTAGCGTCAGTGGGCCGTGGTTACAACCGAGACCGATGTGTGACGTTGCTTGTGGTGGACGATCAAAATACCGACTGGTCCAAGTACTTTAGAGGTCGGAGGCTGCCGGGAGAATGGGACATCCGCGTGGAACAGGCGGAATTCAGGGAACTCTCAGTAACTGCGAACTCGGATGGAGCCAACGTTTCCATGGCGGTGTACCGTAGTGGTACGAAAGTGACACGATGTTTCAAGCCTGACTTCGTGCTGGTGAGGCAAAACGTACGTGACGCTGGCGCGGATCATCGCGCTCTACTGCTTGGACTCAAGTTCGGAGGAGTTCCTTCCATCAACAGCCTCAACTCAATTTACCATTTCCAAGATCGTCCTTGGGTTTTTGGACATCTTCTACAGCTTCAGAGGCGCCTTGGACGGGAGAACTTTCCTCTCATCGAACAAACCTACTACCACAACCACACTGACATGGTGAGTAGTTAA
Protein
MKISSLYLIDNFGRIKSYRCKIVSGYIVCVGFVPPVRVPVWWRGGGARSSVEALRRFSSGDLQSECDEGEVDPYTGRAATTSQPKPQAQKPTSQATGGSLGGSSGPPPAPSAAAAGGDLSLNLRGGSRGTSAPSSPAKSRESLLQRVQSLTGAARDQGANLLGSVTSVASVGRGYNRDRCVTLLVVDDQNTDWSKYFRGRRLPGEWDIRVEQAEFRELSVTANSDGANVSMAVYRSGTKVTRCFKPDFVLVRQNVRDAGADHRALLLGLKFGGVPSINSLNSIYHFQDRPWVFGHLLQLQRRLGRENFPLIEQTYYHNHTDMVSS
Summary
Description
Plays a significant role in nervous system function, which is subtle at the cellular level but manifests itself in complex behavior.
Subunit
Identified in a complex with Syt1 and nwk.
Miscellaneous
Readthrough of the terminator UGA may occur between the codons for 581-Met and 583-Glu.
Similarity
Belongs to the synapsin family.
Keywords
Alternative splicing
Behavior
Cell junction
Complete proteome
Direct protein sequencing
Phosphoprotein
Reference proteome
Synapse
Feature
chain Synapsin
splice variant In isoform C.
splice variant In isoform C.
Uniprot
H9IST1
A0A2W1BLF9
A0A194PYN3
A0A212EZS6
A0A0L7KW36
A0A194R2F0
+ More
A0A154PKA5 A0A158P1M1 A0A139W8P6 A0A0L7R076 A0A2J7RHV3 D7EHT3 E2AKR8 A0A195E2W3 A0A3L8DSW3 A0A067RRB7 E2BNZ5 A0A1W4XJQ1 A0A023F2M3 T1I6V4 A0A2A3EK44 U5ENF5 B3P4K1 B0W8H3 A0A182GE30 E2QCZ0 A0A1Q3FV74 Q24546-3 B4JIL5 E2QCY9 Q5TNR4 A0A3B0K075 A0A0P6B7D4 Q24546 B4NFF4 A0A1A9Y8Y1 A0A1A9V922 A0A1A9ZRQ9 A0A1B0BMG1 A0A2S2R321
A0A154PKA5 A0A158P1M1 A0A139W8P6 A0A0L7R076 A0A2J7RHV3 D7EHT3 E2AKR8 A0A195E2W3 A0A3L8DSW3 A0A067RRB7 E2BNZ5 A0A1W4XJQ1 A0A023F2M3 T1I6V4 A0A2A3EK44 U5ENF5 B3P4K1 B0W8H3 A0A182GE30 E2QCZ0 A0A1Q3FV74 Q24546-3 B4JIL5 E2QCY9 Q5TNR4 A0A3B0K075 A0A0P6B7D4 Q24546 B4NFF4 A0A1A9Y8Y1 A0A1A9V922 A0A1A9ZRQ9 A0A1B0BMG1 A0A2S2R321
Pubmed
EMBL
BABH01013165
BABH01013166
BABH01013167
BABH01013168
KZ150009
PZC75111.1
+ More
KQ459586 KPI97859.1 AGBW02011240 OWR46944.1 JTDY01005000 KOB67492.1 KQ460883 KPJ11425.1 KQ434943 KZC12286.1 ADTU01006617 ADTU01006618 KQ973114 KXZ75661.1 KQ414672 KOC64255.1 NEVH01003737 PNF40415.1 DS497665 EFA12118.1 GL440378 EFN65978.1 KQ979709 KYN19421.1 QOIP01000004 RLU23471.1 KK852511 KDR22299.1 GL449511 EFN82587.1 GBBI01003336 JAC15376.1 ACPB03000880 KZ288231 PBC31566.1 GANO01004128 JAB55743.1 CH954181 EDV49654.1 DS231859 EDS39023.1 JXUM01057246 JXUM01057247 JXUM01057248 KQ561946 KXJ77083.1 AE014297 AAN13463.2 GFDL01003590 JAV31455.1 X95453 X95454 CH916369 EDV93096.1 AAF54506.3 AAAB01008982 EAL39109.4 OUUW01000013 SPP87717.1 GDIP01019128 JAM84587.1 CH964251 EDW83021.2 JXJN01016890 JXJN01016891 GGMS01015244 MBY84447.1
KQ459586 KPI97859.1 AGBW02011240 OWR46944.1 JTDY01005000 KOB67492.1 KQ460883 KPJ11425.1 KQ434943 KZC12286.1 ADTU01006617 ADTU01006618 KQ973114 KXZ75661.1 KQ414672 KOC64255.1 NEVH01003737 PNF40415.1 DS497665 EFA12118.1 GL440378 EFN65978.1 KQ979709 KYN19421.1 QOIP01000004 RLU23471.1 KK852511 KDR22299.1 GL449511 EFN82587.1 GBBI01003336 JAC15376.1 ACPB03000880 KZ288231 PBC31566.1 GANO01004128 JAB55743.1 CH954181 EDV49654.1 DS231859 EDS39023.1 JXUM01057246 JXUM01057247 JXUM01057248 KQ561946 KXJ77083.1 AE014297 AAN13463.2 GFDL01003590 JAV31455.1 X95453 X95454 CH916369 EDV93096.1 AAF54506.3 AAAB01008982 EAL39109.4 OUUW01000013 SPP87717.1 GDIP01019128 JAM84587.1 CH964251 EDW83021.2 JXJN01016890 JXJN01016891 GGMS01015244 MBY84447.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000037510
UP000053240
UP000076502
+ More
UP000005205 UP000007266 UP000053825 UP000235965 UP000000311 UP000078492 UP000279307 UP000027135 UP000008237 UP000192223 UP000015103 UP000242457 UP000008711 UP000002320 UP000069940 UP000249989 UP000000803 UP000001070 UP000007062 UP000268350 UP000007798 UP000092443 UP000078200 UP000092445 UP000092460
UP000005205 UP000007266 UP000053825 UP000235965 UP000000311 UP000078492 UP000279307 UP000027135 UP000008237 UP000192223 UP000015103 UP000242457 UP000008711 UP000002320 UP000069940 UP000249989 UP000000803 UP000001070 UP000007062 UP000268350 UP000007798 UP000092443 UP000078200 UP000092445 UP000092460
Interpro
SUPFAM
SSF52440
SSF52440
Gene 3D
ProteinModelPortal
H9IST1
A0A2W1BLF9
A0A194PYN3
A0A212EZS6
A0A0L7KW36
A0A194R2F0
+ More
A0A154PKA5 A0A158P1M1 A0A139W8P6 A0A0L7R076 A0A2J7RHV3 D7EHT3 E2AKR8 A0A195E2W3 A0A3L8DSW3 A0A067RRB7 E2BNZ5 A0A1W4XJQ1 A0A023F2M3 T1I6V4 A0A2A3EK44 U5ENF5 B3P4K1 B0W8H3 A0A182GE30 E2QCZ0 A0A1Q3FV74 Q24546-3 B4JIL5 E2QCY9 Q5TNR4 A0A3B0K075 A0A0P6B7D4 Q24546 B4NFF4 A0A1A9Y8Y1 A0A1A9V922 A0A1A9ZRQ9 A0A1B0BMG1 A0A2S2R321
A0A154PKA5 A0A158P1M1 A0A139W8P6 A0A0L7R076 A0A2J7RHV3 D7EHT3 E2AKR8 A0A195E2W3 A0A3L8DSW3 A0A067RRB7 E2BNZ5 A0A1W4XJQ1 A0A023F2M3 T1I6V4 A0A2A3EK44 U5ENF5 B3P4K1 B0W8H3 A0A182GE30 E2QCZ0 A0A1Q3FV74 Q24546-3 B4JIL5 E2QCY9 Q5TNR4 A0A3B0K075 A0A0P6B7D4 Q24546 B4NFF4 A0A1A9Y8Y1 A0A1A9V922 A0A1A9ZRQ9 A0A1B0BMG1 A0A2S2R321
PDB
1I7N
E-value=6.86046e-41,
Score=420
Ontologies
GO
PANTHER
Topology
Subcellular location
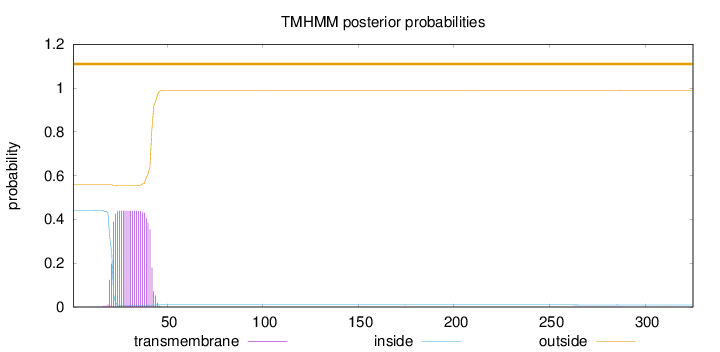
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.22014
Exp number, first 60 AAs:
9.19598
Total prob of N-in:
0.44172
outside
1 - 325
Population Genetic Test Statistics
Pi
307.865287
Theta
193.519713
Tajima's D
1.948218
CLR
0.578757
CSRT
0.870456477176141
Interpretation
Uncertain
