Gene
KWMTBOMO13086 Validated by peptides from experiments
Annotation
PREDICTED:_uncharacterized_protein_LOC101740299_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.125
Sequence
CDS
ATGTCAATCAAGATTAACTTCAACCAGCCTTTCTATGGTGTAGCCCACGCTAGAGAATATAGGACCCCATCATGTATGGCGCTCGGCAATGGCACCGAATCGCTAAACTTTGATATCAACTTGATCGCTCCACAGGGATCGCCTGATCATTGCGGTGTATTCTGGAATAACCGCACTGATGAACGATCGTTGCCTTTGGCGGTTCGGGTGCATCGGACACTCGAGCTGGCCGACGACAAGTTTTACGTCATAACCTGCGGCAAAGCTGGTTTTAAAAACTCTCGGAACGAAACTTCGCTTGTCTCCCTACGAATGCTCGATGACCAAGGACGGAAAGTATTGAACGTTGGCTTCGGAATGCCTTACACGCTGAAAGCCGAAATCAGTAAATCTGATGGAACGCATGGAATCAGATTAAAGAATTGTTTCGGATTCAACATGAAGAATAATACCGCGGAACTACTCGATAATAAAGGGTGCCCGATAAAACAGAGTTCGGTAATAGTAAGGTCTACAGGTGGAACGGCCGAGTTGGCGATAGCAGCGATGTTTCGGTTCCCGGACTCATCTCAAGTTAACTTCCAATGCGACATCGCTATTTGCAAAGGTAGCACTTGTGGAACTGTAGAATGTGATACCGAGGGCAGTGAAAGTCCAGGTGATGAGGAAGGGACCGTGACGGCATCCACGGGAGTGTTCGTTTTAGACCCCAACGACAACACAATCGCAGCCATGTCCTGTATGGAGGGAAACGTGCGGCCATTGTGGCTATTGTATCTGGCCATCGCGCTCGGAGTCATGTTCCTCGTCATGCTACTCATCAATTGCTTCCTTTGTACGGCTATGACGTGTTCGTGTGCAAGGACAGATGTAATTGAGAAAGATCCATCCGTAGTAGAAGATTATGATCCATATCGCAGTTGGCACGGTAGCCAGTACGGCAGCAGGTATCCGTCTTCAACAATTCACTCAACGAGGTCTGTATCGGACAACAGTGATCATTATGCTATAGTGCAGTCAAGGCCGGGCAGTCGCCACTCCGGCATACATCGAAACCGACACTAA
Protein
MSIKINFNQPFYGVAHAREYRTPSCMALGNGTESLNFDINLIAPQGSPDHCGVFWNNRTDERSLPLAVRVHRTLELADDKFYVITCGKAGFKNSRNETSLVSLRMLDDQGRKVLNVGFGMPYTLKAEISKSDGTHGIRLKNCFGFNMKNNTAELLDNKGCPIKQSSVIVRSTGGTAELAIAAMFRFPDSSQVNFQCDIAICKGSTCGTVECDTEGSESPGDEEGTVTASTGVFVLDPNDNTIAAMSCMEGNVRPLWLLYLAIALGVMFLVMLLINCFLCTAMTCSCARTDVIEKDPSVVEDYDPYRSWHGSQYGSRYPSSTIHSTRSVSDNSDHYAIVQSRPGSRHSGIHRNRH
Summary
Uniprot
A0A2W1BJ65
A0A212EZN1
A0A1E1WL50
A0A194PXV6
A0A194R6D6
A0A3S2LYJ4
+ More
A0A2H1V254 A0A2A3EF56 A0A087ZPM6 A0A067REJ5 A0A1S3DVK9 A0A1B6CA23 A0A139W8P8 A0A154PKB2 A0A0L7R057 A0A2S2R6W0 A0A195CSL1 A0A0N0BDD4 A0A195AU63 E0VGX5 A0A195F6N5 E9I907 A0A158P1H5 A0A026X2P5 A0A195E2P3 A0A3L8DUT0 E2BNZ4 E2AKR9 A0A2P8XWZ4 A0A1B6M6T4 A0A1W4XKH9 J3JWK8 A0A1J1HK05 A0A1J1HL37 A0A224XDC3 A0A1Y1MRW6 A0A1Y9HDL6 A0A023ESA5 A0A2M4BNX4 A0A2M3ZED8 H9IT13 A0A1Q3FPS7 A0A1S4FZH1 A0A1Q3FPV3 A0A2M4CXZ4 A0A2M4BNG7 A0A1I8PVH5 A0A2M4CXZ1 A0A2M4CY47 A0A2M4CZA4 A0A2M4A811 B3LW13 A0A0K8VCZ0 N6SYV2 A0A1I8PVJ3 B0W8H2 A0A0P8XWZ3 A0A0A1XTB8 A0A0Q9X797 B4NFF3 Q9VH17 B4PKM2 A0A232FJ80 A0A1I8MDJ8 B4LWD5 A0A1W4UM02 A0A0K8SR89 Q29AK4 A0A0Q9XAE0 A0A0Q9WYL1 A0A0B4KFK4 A0A0A9WFE9 A0A0Q9WDY1 A0A0L0CGK4 A0A151X7C8 W8BEZ1 A0A1A9Y606 A0A2J7RHV5 A0A0A1WJ44 A0A1A9UX45 A0A0K8VNW8 A0A0R3NFZ5 A0A034W119 B4K5I1 B4G6G2 A0A3B0KH07 T1PC47 W8AUA9 A0A0Q9WED0 B4JIL4 A0A0A1XCE1 A0A1W4U8Z6 A0A0M4EFQ4 I5ANB9 A0A0N8P187 B3P4K2 A0A0R3NL20 A0A0B4KFP1 A0A1L8E3P1
A0A2H1V254 A0A2A3EF56 A0A087ZPM6 A0A067REJ5 A0A1S3DVK9 A0A1B6CA23 A0A139W8P8 A0A154PKB2 A0A0L7R057 A0A2S2R6W0 A0A195CSL1 A0A0N0BDD4 A0A195AU63 E0VGX5 A0A195F6N5 E9I907 A0A158P1H5 A0A026X2P5 A0A195E2P3 A0A3L8DUT0 E2BNZ4 E2AKR9 A0A2P8XWZ4 A0A1B6M6T4 A0A1W4XKH9 J3JWK8 A0A1J1HK05 A0A1J1HL37 A0A224XDC3 A0A1Y1MRW6 A0A1Y9HDL6 A0A023ESA5 A0A2M4BNX4 A0A2M3ZED8 H9IT13 A0A1Q3FPS7 A0A1S4FZH1 A0A1Q3FPV3 A0A2M4CXZ4 A0A2M4BNG7 A0A1I8PVH5 A0A2M4CXZ1 A0A2M4CY47 A0A2M4CZA4 A0A2M4A811 B3LW13 A0A0K8VCZ0 N6SYV2 A0A1I8PVJ3 B0W8H2 A0A0P8XWZ3 A0A0A1XTB8 A0A0Q9X797 B4NFF3 Q9VH17 B4PKM2 A0A232FJ80 A0A1I8MDJ8 B4LWD5 A0A1W4UM02 A0A0K8SR89 Q29AK4 A0A0Q9XAE0 A0A0Q9WYL1 A0A0B4KFK4 A0A0A9WFE9 A0A0Q9WDY1 A0A0L0CGK4 A0A151X7C8 W8BEZ1 A0A1A9Y606 A0A2J7RHV5 A0A0A1WJ44 A0A1A9UX45 A0A0K8VNW8 A0A0R3NFZ5 A0A034W119 B4K5I1 B4G6G2 A0A3B0KH07 T1PC47 W8AUA9 A0A0Q9WED0 B4JIL4 A0A0A1XCE1 A0A1W4U8Z6 A0A0M4EFQ4 I5ANB9 A0A0N8P187 B3P4K2 A0A0R3NL20 A0A0B4KFP1 A0A1L8E3P1
Pubmed
28756777
22118469
26354079
24845553
18362917
19820115
+ More
20566863 21282665 21347285 24508170 30249741 20798317 29403074 22516182 23537049 28004739 24945155 19121390 17994087 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 28648823 25315136 26823975 15632085 18057021 25401762 26108605 24495485 25348373
20566863 21282665 21347285 24508170 30249741 20798317 29403074 22516182 23537049 28004739 24945155 19121390 17994087 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 28648823 25315136 26823975 15632085 18057021 25401762 26108605 24495485 25348373
EMBL
KZ150009
PZC75112.1
AGBW02011240
OWR46943.1
GDQN01003301
JAT87753.1
+ More
KQ459586 KPI97858.1 KQ460883 KPJ11426.1 RSAL01000111 RVE47098.1 ODYU01000339 SOQ34925.1 KZ288262 PBC30348.1 KK852511 KDR22301.1 GEDC01026975 JAS10323.1 KQ973114 KXZ75662.1 KQ434943 KZC12285.1 KQ414672 KOC64254.1 GGMS01016491 MBY85694.1 KQ977305 KYN03698.1 KQ435878 KOX69941.1 KQ976741 KYM75597.1 DS235152 EEB12631.1 KQ981756 KYN36123.1 GL761667 EFZ22951.1 ADTU01006619 ADTU01006620 ADTU01006621 ADTU01006622 KK107024 EZA62286.1 KQ979709 KYN19418.1 QOIP01000004 RLU24052.1 GL449511 EFN82586.1 GL440378 EFN65979.1 PYGN01001223 PSN36518.1 GEBQ01008349 JAT31628.1 BT127626 KB632056 AEE62588.1 ERL88354.1 CVRI01000006 CRK88251.1 CRK88250.1 GFTR01006009 JAW10417.1 GEZM01023451 JAV88381.1 GAPW01001471 JAC12127.1 GGFJ01005563 MBW54704.1 GGFM01006196 MBW26947.1 BABH01013175 BABH01013176 GFDL01005557 JAV29488.1 GFDL01005542 JAV29503.1 GGFL01006024 MBW70202.1 GGFJ01005488 MBW54629.1 GGFL01006025 MBW70203.1 GGFL01006011 MBW70189.1 GGFL01006010 MBW70188.1 GGFK01003615 MBW36936.1 CH902617 EDV41546.2 GDHF01021717 GDHF01015505 GDHF01014437 JAI30597.1 JAI36809.1 JAI37877.1 APGK01059103 APGK01059104 APGK01059105 APGK01059106 KB741292 ENN70413.1 DS231859 EDS39022.1 KPU79235.1 GBXI01000439 JAD13853.1 CH933806 KRG00802.1 CH964251 EDW83020.2 AE014297 AAF54504.4 CM000160 EDW96781.2 NNAY01000126 OXU30732.1 CH940650 EDW66572.2 GBRD01010088 GDHC01003718 GDHC01001779 JAG55736.1 JAQ14911.1 JAQ16850.1 CM000070 EAL27345.3 KRG00803.1 KRG00806.1 KRG00805.1 AGB95836.1 GBHO01037503 GBHO01000705 GBHO01000704 JAG06101.1 JAG42899.1 JAG42900.1 KRF82808.1 JRES01000409 KNC31538.1 KQ982450 KYQ56239.1 GAMC01018263 GAMC01018261 JAB88294.1 NEVH01003737 PNF40413.1 GBXI01015747 JAC98544.1 GDHF01015226 GDHF01012406 GDHF01012089 JAI37088.1 JAI39908.1 JAI40225.1 KRS99965.1 GAKP01010573 GAKP01010571 JAC48379.1 EDW14018.2 CH479179 EDW23881.1 OUUW01000013 SPP87720.1 KA645508 AFP60137.1 GAMC01018262 JAB88293.1 KRF82810.1 CH916369 EDV93095.1 GBXI01006164 JAD08128.1 CP012526 ALC47041.1 EIM52454.2 KPU79236.1 CH954181 EDV49655.1 KRS99964.1 AGB95835.1 GFDF01000731 JAV13353.1
KQ459586 KPI97858.1 KQ460883 KPJ11426.1 RSAL01000111 RVE47098.1 ODYU01000339 SOQ34925.1 KZ288262 PBC30348.1 KK852511 KDR22301.1 GEDC01026975 JAS10323.1 KQ973114 KXZ75662.1 KQ434943 KZC12285.1 KQ414672 KOC64254.1 GGMS01016491 MBY85694.1 KQ977305 KYN03698.1 KQ435878 KOX69941.1 KQ976741 KYM75597.1 DS235152 EEB12631.1 KQ981756 KYN36123.1 GL761667 EFZ22951.1 ADTU01006619 ADTU01006620 ADTU01006621 ADTU01006622 KK107024 EZA62286.1 KQ979709 KYN19418.1 QOIP01000004 RLU24052.1 GL449511 EFN82586.1 GL440378 EFN65979.1 PYGN01001223 PSN36518.1 GEBQ01008349 JAT31628.1 BT127626 KB632056 AEE62588.1 ERL88354.1 CVRI01000006 CRK88251.1 CRK88250.1 GFTR01006009 JAW10417.1 GEZM01023451 JAV88381.1 GAPW01001471 JAC12127.1 GGFJ01005563 MBW54704.1 GGFM01006196 MBW26947.1 BABH01013175 BABH01013176 GFDL01005557 JAV29488.1 GFDL01005542 JAV29503.1 GGFL01006024 MBW70202.1 GGFJ01005488 MBW54629.1 GGFL01006025 MBW70203.1 GGFL01006011 MBW70189.1 GGFL01006010 MBW70188.1 GGFK01003615 MBW36936.1 CH902617 EDV41546.2 GDHF01021717 GDHF01015505 GDHF01014437 JAI30597.1 JAI36809.1 JAI37877.1 APGK01059103 APGK01059104 APGK01059105 APGK01059106 KB741292 ENN70413.1 DS231859 EDS39022.1 KPU79235.1 GBXI01000439 JAD13853.1 CH933806 KRG00802.1 CH964251 EDW83020.2 AE014297 AAF54504.4 CM000160 EDW96781.2 NNAY01000126 OXU30732.1 CH940650 EDW66572.2 GBRD01010088 GDHC01003718 GDHC01001779 JAG55736.1 JAQ14911.1 JAQ16850.1 CM000070 EAL27345.3 KRG00803.1 KRG00806.1 KRG00805.1 AGB95836.1 GBHO01037503 GBHO01000705 GBHO01000704 JAG06101.1 JAG42899.1 JAG42900.1 KRF82808.1 JRES01000409 KNC31538.1 KQ982450 KYQ56239.1 GAMC01018263 GAMC01018261 JAB88294.1 NEVH01003737 PNF40413.1 GBXI01015747 JAC98544.1 GDHF01015226 GDHF01012406 GDHF01012089 JAI37088.1 JAI39908.1 JAI40225.1 KRS99965.1 GAKP01010573 GAKP01010571 JAC48379.1 EDW14018.2 CH479179 EDW23881.1 OUUW01000013 SPP87720.1 KA645508 AFP60137.1 GAMC01018262 JAB88293.1 KRF82810.1 CH916369 EDV93095.1 GBXI01006164 JAD08128.1 CP012526 ALC47041.1 EIM52454.2 KPU79236.1 CH954181 EDV49655.1 KRS99964.1 AGB95835.1 GFDF01000731 JAV13353.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000283053
UP000242457
UP000005203
+ More
UP000027135 UP000079169 UP000007266 UP000076502 UP000053825 UP000078542 UP000053105 UP000078540 UP000009046 UP000078541 UP000005205 UP000053097 UP000078492 UP000279307 UP000008237 UP000000311 UP000245037 UP000192223 UP000030742 UP000183832 UP000075900 UP000005204 UP000095300 UP000007801 UP000019118 UP000002320 UP000009192 UP000007798 UP000000803 UP000002282 UP000215335 UP000095301 UP000008792 UP000192221 UP000001819 UP000037069 UP000075809 UP000092443 UP000235965 UP000078200 UP000008744 UP000268350 UP000001070 UP000092553 UP000008711
UP000027135 UP000079169 UP000007266 UP000076502 UP000053825 UP000078542 UP000053105 UP000078540 UP000009046 UP000078541 UP000005205 UP000053097 UP000078492 UP000279307 UP000008237 UP000000311 UP000245037 UP000192223 UP000030742 UP000183832 UP000075900 UP000005204 UP000095300 UP000007801 UP000019118 UP000002320 UP000009192 UP000007798 UP000000803 UP000002282 UP000215335 UP000095301 UP000008792 UP000192221 UP000001819 UP000037069 UP000075809 UP000092443 UP000235965 UP000078200 UP000008744 UP000268350 UP000001070 UP000092553 UP000008711
Pfam
PF00100 Zona_pellucida
Interpro
IPR001507
ZP_dom
ProteinModelPortal
A0A2W1BJ65
A0A212EZN1
A0A1E1WL50
A0A194PXV6
A0A194R6D6
A0A3S2LYJ4
+ More
A0A2H1V254 A0A2A3EF56 A0A087ZPM6 A0A067REJ5 A0A1S3DVK9 A0A1B6CA23 A0A139W8P8 A0A154PKB2 A0A0L7R057 A0A2S2R6W0 A0A195CSL1 A0A0N0BDD4 A0A195AU63 E0VGX5 A0A195F6N5 E9I907 A0A158P1H5 A0A026X2P5 A0A195E2P3 A0A3L8DUT0 E2BNZ4 E2AKR9 A0A2P8XWZ4 A0A1B6M6T4 A0A1W4XKH9 J3JWK8 A0A1J1HK05 A0A1J1HL37 A0A224XDC3 A0A1Y1MRW6 A0A1Y9HDL6 A0A023ESA5 A0A2M4BNX4 A0A2M3ZED8 H9IT13 A0A1Q3FPS7 A0A1S4FZH1 A0A1Q3FPV3 A0A2M4CXZ4 A0A2M4BNG7 A0A1I8PVH5 A0A2M4CXZ1 A0A2M4CY47 A0A2M4CZA4 A0A2M4A811 B3LW13 A0A0K8VCZ0 N6SYV2 A0A1I8PVJ3 B0W8H2 A0A0P8XWZ3 A0A0A1XTB8 A0A0Q9X797 B4NFF3 Q9VH17 B4PKM2 A0A232FJ80 A0A1I8MDJ8 B4LWD5 A0A1W4UM02 A0A0K8SR89 Q29AK4 A0A0Q9XAE0 A0A0Q9WYL1 A0A0B4KFK4 A0A0A9WFE9 A0A0Q9WDY1 A0A0L0CGK4 A0A151X7C8 W8BEZ1 A0A1A9Y606 A0A2J7RHV5 A0A0A1WJ44 A0A1A9UX45 A0A0K8VNW8 A0A0R3NFZ5 A0A034W119 B4K5I1 B4G6G2 A0A3B0KH07 T1PC47 W8AUA9 A0A0Q9WED0 B4JIL4 A0A0A1XCE1 A0A1W4U8Z6 A0A0M4EFQ4 I5ANB9 A0A0N8P187 B3P4K2 A0A0R3NL20 A0A0B4KFP1 A0A1L8E3P1
A0A2H1V254 A0A2A3EF56 A0A087ZPM6 A0A067REJ5 A0A1S3DVK9 A0A1B6CA23 A0A139W8P8 A0A154PKB2 A0A0L7R057 A0A2S2R6W0 A0A195CSL1 A0A0N0BDD4 A0A195AU63 E0VGX5 A0A195F6N5 E9I907 A0A158P1H5 A0A026X2P5 A0A195E2P3 A0A3L8DUT0 E2BNZ4 E2AKR9 A0A2P8XWZ4 A0A1B6M6T4 A0A1W4XKH9 J3JWK8 A0A1J1HK05 A0A1J1HL37 A0A224XDC3 A0A1Y1MRW6 A0A1Y9HDL6 A0A023ESA5 A0A2M4BNX4 A0A2M3ZED8 H9IT13 A0A1Q3FPS7 A0A1S4FZH1 A0A1Q3FPV3 A0A2M4CXZ4 A0A2M4BNG7 A0A1I8PVH5 A0A2M4CXZ1 A0A2M4CY47 A0A2M4CZA4 A0A2M4A811 B3LW13 A0A0K8VCZ0 N6SYV2 A0A1I8PVJ3 B0W8H2 A0A0P8XWZ3 A0A0A1XTB8 A0A0Q9X797 B4NFF3 Q9VH17 B4PKM2 A0A232FJ80 A0A1I8MDJ8 B4LWD5 A0A1W4UM02 A0A0K8SR89 Q29AK4 A0A0Q9XAE0 A0A0Q9WYL1 A0A0B4KFK4 A0A0A9WFE9 A0A0Q9WDY1 A0A0L0CGK4 A0A151X7C8 W8BEZ1 A0A1A9Y606 A0A2J7RHV5 A0A0A1WJ44 A0A1A9UX45 A0A0K8VNW8 A0A0R3NFZ5 A0A034W119 B4K5I1 B4G6G2 A0A3B0KH07 T1PC47 W8AUA9 A0A0Q9WED0 B4JIL4 A0A0A1XCE1 A0A1W4U8Z6 A0A0M4EFQ4 I5ANB9 A0A0N8P187 B3P4K2 A0A0R3NL20 A0A0B4KFP1 A0A1L8E3P1
Ontologies
GO
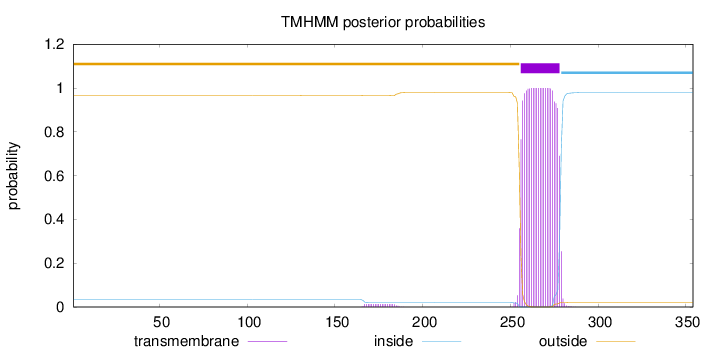
Topology
Length:
354
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.15265
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.03443
outside
1 - 255
TMhelix
256 - 278
inside
279 - 354
Population Genetic Test Statistics
Pi
284.441391
Theta
197.772712
Tajima's D
1.499103
CLR
0.186314
CSRT
0.789460526973651
Interpretation
Uncertain
