Gene
KWMTBOMO13084 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000307
Annotation
PREDICTED:_carboxypeptidase_E-like_[Bombyx_mori]
Full name
Tetraspanin
Location in the cell
Extracellular Reliability : 2.757
Sequence
CDS
ATGGCTTTGTATAGTTTCGTGTGTTTTTCTCTTTTATTAACGGTGTCCGCAGAGTTCCAGTGGAAACATCATAACAATGAAGAACTACCCTTGGTTTTACAAGAAGTACATAATAACTGCCCGAATATAACTAGAATCTATGCATTAAGTGAGCCCTCAGTTTGCAATGTTCCCTTGTATGTCATAGAATTTGCACAGGTTCCTGGTTTTCATCGACCGTATATACCAGAAGCCAAATATATAGGGAATATTCACGGAAACGAAGTTCTGGGTCGAGAGCTGCTTTTGGGATTGGCACATTATCTTTGTGATCAATATAGGAAAAATGATCCTGAAATAAAAGCTCTGATTACTAATACCAGAATCCATTTGTTACCATCAATGAATCCTGATGGATGGCAACTTGCCACTGATACAGGTGGAAAAGATTACTTGATAGGCCGAACAAATAATCATGAAGTGGATCTCAACAGAAATTTCCCAGACTTAGATGCTATTACATTTGACTTTGAACGCCAGGGTCTTAGCCACAACAACCATTTATTGAAGGATCTCACTCAATTGAGTGCTCCTCTGGAGCCAGAAACAAGAGCTGTTATGCGCTGGATTATGTCTACACCATTTGTATTGAGTGCTGCAATACATGGAGGTGACTTGGTTGCTAATTACCCATATGATGAGAGCAAAACTGGAGCTTCGGCCGCAGAGTATTCTGCAAGTCCAGATGATGAAACCTTCAAGGAATTGGCAATGACTTATGCATTGGCCCACGCGGACATGGCGTCGCCGACCCGTAGGGGATGCCATACCACTAGTAGTGACGATGTCTCATACAACTTCGGCAAACAGGGCGGCGTGACCAATGGAGCGGCTTGGTATAGTTTAAAAGGAGGTAAGCAGTAA
Protein
MALYSFVCFSLLLTVSAEFQWKHHNNEELPLVLQEVHNNCPNITRIYALSEPSVCNVPLYVIEFAQVPGFHRPYIPEAKYIGNIHGNEVLGRELLLGLAHYLCDQYRKNDPEIKALITNTRIHLLPSMNPDGWQLATDTGGKDYLIGRTNNHEVDLNRNFPDLDAITFDFERQGLSHNNHLLKDLTQLSAPLEPETRAVMRWIMSTPFVLSAAIHGGDLVANYPYDESKTGASAAEYSASPDDETFKELAMTYALAHADMASPTRRGCHTTSSDDVSYNFGKQGGVTNGAAWYSLKGGKQ
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9IST0
A0A2H1V3R4
A0A2W1BTP9
A0A212F4B5
A0A194PX08
A0A0L7LI35
+ More
A0A139WAF4 V5I9C3 A0A1W4X774 A0A1Y1KLK3 A0A1I9WL51 A0A0T6AVF3 T1HGI1 A0A1V1FKQ5 A0A2J7RN85 A0A067QWF9 A0A1B6CEM3 A0A1B6FQI7 E0W2S8 A0A0A9XL37 U4UJ45 A0A226DM88 A0A2H8TIL6 A0A2R7VX82 A0A0P5A0I0 A0A0P5ENK7 A0A1D2NM88 E9FTT2 A0A2P8ZDS6 A0A0P5E1S9 A0A2S2NLB9 J9JM00 A0A0P4ZGN0 A0A0P5BQN4 A0A0P6HJT4 A0A0P5SXF1 A0A0P5E6J9 A0A0P5E828 A0A0P5CH61 A0A0N8A7Q2 A0A224XI08 A0A0P5YKZ9 T1IXL0 T1IMV6 N6T861 A0A194R146 A0A0V1K0G9 A0A0V1FV88 A0A0V1K1P9 A0A0V0YNU5 A0A0V1IVZ7 A0A1W0WG09 A0A0V1I4E1 A0A0V0YPH4 A0A0V1I4G2 A0A0V1MVC2 A0A0V1CRW1 A0A0V0U0G8 A0A0V1K0S8 A0A0V0VQ75 A0A0V1CRX2 A0A0V1A5R3 A0A0V0RX02 A0A0V0VQ80 A0A0V1IVX8 A0A0V1CS93 A0A0V0RX28 A0A0V1BK60 A0A0V1BKU9 T1L1W8 A0A0V1BKD1 A0A1S5QN46 A0A0B1PN22 A0A087TJM9 A0A0V1I4S6 A0A0V1MVA7 A0A077Z5G2 A0A0V1I5I9 A0A0V1L7P5 A0A0V0WZI9 A0A1D1UKZ8 A0A3S3QWN2 V4BAS9 A0A0V1BKN8 A0A0V1BK39 A0A0V1P2P8 A0A0V1L7N3 A0A0V1BKN0 A0A0V1L7S7 A0A0V0WZ87 A0A0U4BW51 A0A2T7PX17 A0A1S3JWZ2 A0A3Q2YPC6 A0A1Y3EH44 A0A3Q3MD33 A0A3S1BQA2 A0A3B4WNY2 A0A2I4C6T6
A0A139WAF4 V5I9C3 A0A1W4X774 A0A1Y1KLK3 A0A1I9WL51 A0A0T6AVF3 T1HGI1 A0A1V1FKQ5 A0A2J7RN85 A0A067QWF9 A0A1B6CEM3 A0A1B6FQI7 E0W2S8 A0A0A9XL37 U4UJ45 A0A226DM88 A0A2H8TIL6 A0A2R7VX82 A0A0P5A0I0 A0A0P5ENK7 A0A1D2NM88 E9FTT2 A0A2P8ZDS6 A0A0P5E1S9 A0A2S2NLB9 J9JM00 A0A0P4ZGN0 A0A0P5BQN4 A0A0P6HJT4 A0A0P5SXF1 A0A0P5E6J9 A0A0P5E828 A0A0P5CH61 A0A0N8A7Q2 A0A224XI08 A0A0P5YKZ9 T1IXL0 T1IMV6 N6T861 A0A194R146 A0A0V1K0G9 A0A0V1FV88 A0A0V1K1P9 A0A0V0YNU5 A0A0V1IVZ7 A0A1W0WG09 A0A0V1I4E1 A0A0V0YPH4 A0A0V1I4G2 A0A0V1MVC2 A0A0V1CRW1 A0A0V0U0G8 A0A0V1K0S8 A0A0V0VQ75 A0A0V1CRX2 A0A0V1A5R3 A0A0V0RX02 A0A0V0VQ80 A0A0V1IVX8 A0A0V1CS93 A0A0V0RX28 A0A0V1BK60 A0A0V1BKU9 T1L1W8 A0A0V1BKD1 A0A1S5QN46 A0A0B1PN22 A0A087TJM9 A0A0V1I4S6 A0A0V1MVA7 A0A077Z5G2 A0A0V1I5I9 A0A0V1L7P5 A0A0V0WZI9 A0A1D1UKZ8 A0A3S3QWN2 V4BAS9 A0A0V1BKN8 A0A0V1BK39 A0A0V1P2P8 A0A0V1L7N3 A0A0V1BKN0 A0A0V1L7S7 A0A0V0WZ87 A0A0U4BW51 A0A2T7PX17 A0A1S3JWZ2 A0A3Q2YPC6 A0A1Y3EH44 A0A3Q3MD33 A0A3S1BQA2 A0A3B4WNY2 A0A2I4C6T6
Pubmed
EMBL
BABH01013177
ODYU01000339
SOQ34924.1
KZ150009
PZC75113.1
AGBW02010414
+ More
OWR48562.1 KQ459586 KPI97857.1 JTDY01001037 KOB75065.1 KQ971409 KYB24878.1 GALX01003185 JAB65281.1 GEZM01083387 JAV61120.1 KU932235 APA33871.1 LJIG01022706 KRT79135.1 ACPB03018778 FX985399 BAX07412.1 NEVH01002545 PNF42294.1 KK853170 KDR10344.1 GEDC01025439 GEDC01007084 JAS11859.1 JAS30214.1 GECZ01017329 JAS52440.1 DS235879 EEB19934.1 GBHO01022965 GBRD01000314 GDHC01003988 GDHC01003389 JAG20639.1 JAG65507.1 JAQ14641.1 JAQ15240.1 KB632379 ERL94094.1 LNIX01000015 OXA46642.1 GFXV01002178 MBW13983.1 KK854143 PTY12072.1 GDIP01210079 JAJ13323.1 GDIP01145436 JAJ77966.1 LJIJ01000006 ODN06398.1 GL732524 EFX89423.1 PYGN01000086 PSN54662.1 GDIP01148235 JAJ75167.1 GGMR01005360 MBY17979.1 ABLF02016878 ABLF02035270 ABLF02062804 GDIP01213083 JAJ10319.1 GDIP01185988 LRGB01003123 JAJ37414.1 KZS04404.1 GDIQ01026810 JAN67927.1 GDIP01138832 JAL64882.1 GDIP01150888 JAJ72514.1 GDIP01145437 JAJ77965.1 GDIP01174322 JAJ49080.1 GDIP01174321 JAJ49081.1 GFTR01008747 JAW07679.1 GDIP01056442 JAM47273.1 AFFK01020101 JH431096 APGK01040397 KB740979 ENN76419.1 KQ460883 KPJ11427.1 JYDV01000025 KRZ40743.1 JYDT01000026 JYDS01000078 KRY89956.1 KRZ26927.1 KRZ40741.1 JYDU01000001 KRY01998.1 KRZ26928.1 MTYJ01000109 OQV14136.1 JYDP01000005 KRZ17758.1 KRY01997.1 KRZ40744.1 KRZ17761.1 JYDO01000035 KRZ75730.1 JYDI01000113 KRY52026.1 JYDJ01000090 KRX44728.1 KRZ40742.1 JYDN01000015 KRX65662.1 KRY52025.1 JYDQ01000031 KRY19806.1 JYDL01000065 KRX19009.1 KRX65663.1 KRZ26926.1 KRY52024.1 KRX19010.1 JYDH01000034 KRY37392.1 KRY37391.1 CAEY01000922 KRY37393.1 KR068529 AMO02546.1 KN538416 KHJ41512.1 KK115515 KFM65318.1 KRZ17759.1 KRZ75729.1 HG805934 CDW55064.1 KRZ17760.1 JYDW01000115 KRZ55429.1 JYDK01000034 KRX81101.1 BDGG01000001 GAU87947.1 NCKU01000630 NCKU01000525 RWS14749.1 RWS15185.1 KB199676 ESP04631.1 KRY37390.1 KRY37389.1 JYDM01000056 KRZ90467.1 KRZ55430.1 KRY37388.1 KRZ55431.1 KRX81100.1 KU218671 ALX00051.1 PZQS01000001 PVD37965.1 LVZM01015902 OUC43146.1 RQTK01000149 RUS86156.1
OWR48562.1 KQ459586 KPI97857.1 JTDY01001037 KOB75065.1 KQ971409 KYB24878.1 GALX01003185 JAB65281.1 GEZM01083387 JAV61120.1 KU932235 APA33871.1 LJIG01022706 KRT79135.1 ACPB03018778 FX985399 BAX07412.1 NEVH01002545 PNF42294.1 KK853170 KDR10344.1 GEDC01025439 GEDC01007084 JAS11859.1 JAS30214.1 GECZ01017329 JAS52440.1 DS235879 EEB19934.1 GBHO01022965 GBRD01000314 GDHC01003988 GDHC01003389 JAG20639.1 JAG65507.1 JAQ14641.1 JAQ15240.1 KB632379 ERL94094.1 LNIX01000015 OXA46642.1 GFXV01002178 MBW13983.1 KK854143 PTY12072.1 GDIP01210079 JAJ13323.1 GDIP01145436 JAJ77966.1 LJIJ01000006 ODN06398.1 GL732524 EFX89423.1 PYGN01000086 PSN54662.1 GDIP01148235 JAJ75167.1 GGMR01005360 MBY17979.1 ABLF02016878 ABLF02035270 ABLF02062804 GDIP01213083 JAJ10319.1 GDIP01185988 LRGB01003123 JAJ37414.1 KZS04404.1 GDIQ01026810 JAN67927.1 GDIP01138832 JAL64882.1 GDIP01150888 JAJ72514.1 GDIP01145437 JAJ77965.1 GDIP01174322 JAJ49080.1 GDIP01174321 JAJ49081.1 GFTR01008747 JAW07679.1 GDIP01056442 JAM47273.1 AFFK01020101 JH431096 APGK01040397 KB740979 ENN76419.1 KQ460883 KPJ11427.1 JYDV01000025 KRZ40743.1 JYDT01000026 JYDS01000078 KRY89956.1 KRZ26927.1 KRZ40741.1 JYDU01000001 KRY01998.1 KRZ26928.1 MTYJ01000109 OQV14136.1 JYDP01000005 KRZ17758.1 KRY01997.1 KRZ40744.1 KRZ17761.1 JYDO01000035 KRZ75730.1 JYDI01000113 KRY52026.1 JYDJ01000090 KRX44728.1 KRZ40742.1 JYDN01000015 KRX65662.1 KRY52025.1 JYDQ01000031 KRY19806.1 JYDL01000065 KRX19009.1 KRX65663.1 KRZ26926.1 KRY52024.1 KRX19010.1 JYDH01000034 KRY37392.1 KRY37391.1 CAEY01000922 KRY37393.1 KR068529 AMO02546.1 KN538416 KHJ41512.1 KK115515 KFM65318.1 KRZ17759.1 KRZ75729.1 HG805934 CDW55064.1 KRZ17760.1 JYDW01000115 KRZ55429.1 JYDK01000034 KRX81101.1 BDGG01000001 GAU87947.1 NCKU01000630 NCKU01000525 RWS14749.1 RWS15185.1 KB199676 ESP04631.1 KRY37390.1 KRY37389.1 JYDM01000056 KRZ90467.1 KRZ55430.1 KRY37388.1 KRZ55431.1 KRX81100.1 KU218671 ALX00051.1 PZQS01000001 PVD37965.1 LVZM01015902 OUC43146.1 RQTK01000149 RUS86156.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000037510
UP000007266
UP000192223
+ More
UP000015103 UP000235965 UP000027135 UP000009046 UP000030742 UP000198287 UP000094527 UP000000305 UP000245037 UP000007819 UP000076858 UP000019118 UP000053240 UP000054826 UP000054805 UP000054995 UP000054815 UP000055024 UP000054843 UP000054653 UP000055048 UP000054681 UP000054783 UP000054630 UP000054776 UP000015104 UP000054359 UP000030665 UP000054721 UP000054673 UP000186922 UP000285301 UP000030746 UP000054924 UP000245119 UP000085678 UP000264820 UP000243006 UP000261640 UP000271974 UP000261360 UP000192220
UP000015103 UP000235965 UP000027135 UP000009046 UP000030742 UP000198287 UP000094527 UP000000305 UP000245037 UP000007819 UP000076858 UP000019118 UP000053240 UP000054826 UP000054805 UP000054995 UP000054815 UP000055024 UP000054843 UP000054653 UP000055048 UP000054681 UP000054783 UP000054630 UP000054776 UP000015104 UP000054359 UP000030665 UP000054721 UP000054673 UP000186922 UP000285301 UP000030746 UP000054924 UP000245119 UP000085678 UP000264820 UP000243006 UP000261640 UP000271974 UP000261360 UP000192220
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IST0
A0A2H1V3R4
A0A2W1BTP9
A0A212F4B5
A0A194PX08
A0A0L7LI35
+ More
A0A139WAF4 V5I9C3 A0A1W4X774 A0A1Y1KLK3 A0A1I9WL51 A0A0T6AVF3 T1HGI1 A0A1V1FKQ5 A0A2J7RN85 A0A067QWF9 A0A1B6CEM3 A0A1B6FQI7 E0W2S8 A0A0A9XL37 U4UJ45 A0A226DM88 A0A2H8TIL6 A0A2R7VX82 A0A0P5A0I0 A0A0P5ENK7 A0A1D2NM88 E9FTT2 A0A2P8ZDS6 A0A0P5E1S9 A0A2S2NLB9 J9JM00 A0A0P4ZGN0 A0A0P5BQN4 A0A0P6HJT4 A0A0P5SXF1 A0A0P5E6J9 A0A0P5E828 A0A0P5CH61 A0A0N8A7Q2 A0A224XI08 A0A0P5YKZ9 T1IXL0 T1IMV6 N6T861 A0A194R146 A0A0V1K0G9 A0A0V1FV88 A0A0V1K1P9 A0A0V0YNU5 A0A0V1IVZ7 A0A1W0WG09 A0A0V1I4E1 A0A0V0YPH4 A0A0V1I4G2 A0A0V1MVC2 A0A0V1CRW1 A0A0V0U0G8 A0A0V1K0S8 A0A0V0VQ75 A0A0V1CRX2 A0A0V1A5R3 A0A0V0RX02 A0A0V0VQ80 A0A0V1IVX8 A0A0V1CS93 A0A0V0RX28 A0A0V1BK60 A0A0V1BKU9 T1L1W8 A0A0V1BKD1 A0A1S5QN46 A0A0B1PN22 A0A087TJM9 A0A0V1I4S6 A0A0V1MVA7 A0A077Z5G2 A0A0V1I5I9 A0A0V1L7P5 A0A0V0WZI9 A0A1D1UKZ8 A0A3S3QWN2 V4BAS9 A0A0V1BKN8 A0A0V1BK39 A0A0V1P2P8 A0A0V1L7N3 A0A0V1BKN0 A0A0V1L7S7 A0A0V0WZ87 A0A0U4BW51 A0A2T7PX17 A0A1S3JWZ2 A0A3Q2YPC6 A0A1Y3EH44 A0A3Q3MD33 A0A3S1BQA2 A0A3B4WNY2 A0A2I4C6T6
A0A139WAF4 V5I9C3 A0A1W4X774 A0A1Y1KLK3 A0A1I9WL51 A0A0T6AVF3 T1HGI1 A0A1V1FKQ5 A0A2J7RN85 A0A067QWF9 A0A1B6CEM3 A0A1B6FQI7 E0W2S8 A0A0A9XL37 U4UJ45 A0A226DM88 A0A2H8TIL6 A0A2R7VX82 A0A0P5A0I0 A0A0P5ENK7 A0A1D2NM88 E9FTT2 A0A2P8ZDS6 A0A0P5E1S9 A0A2S2NLB9 J9JM00 A0A0P4ZGN0 A0A0P5BQN4 A0A0P6HJT4 A0A0P5SXF1 A0A0P5E6J9 A0A0P5E828 A0A0P5CH61 A0A0N8A7Q2 A0A224XI08 A0A0P5YKZ9 T1IXL0 T1IMV6 N6T861 A0A194R146 A0A0V1K0G9 A0A0V1FV88 A0A0V1K1P9 A0A0V0YNU5 A0A0V1IVZ7 A0A1W0WG09 A0A0V1I4E1 A0A0V0YPH4 A0A0V1I4G2 A0A0V1MVC2 A0A0V1CRW1 A0A0V0U0G8 A0A0V1K0S8 A0A0V0VQ75 A0A0V1CRX2 A0A0V1A5R3 A0A0V0RX02 A0A0V0VQ80 A0A0V1IVX8 A0A0V1CS93 A0A0V0RX28 A0A0V1BK60 A0A0V1BKU9 T1L1W8 A0A0V1BKD1 A0A1S5QN46 A0A0B1PN22 A0A087TJM9 A0A0V1I4S6 A0A0V1MVA7 A0A077Z5G2 A0A0V1I5I9 A0A0V1L7P5 A0A0V0WZI9 A0A1D1UKZ8 A0A3S3QWN2 V4BAS9 A0A0V1BKN8 A0A0V1BK39 A0A0V1P2P8 A0A0V1L7N3 A0A0V1BKN0 A0A0V1L7S7 A0A0V0WZ87 A0A0U4BW51 A0A2T7PX17 A0A1S3JWZ2 A0A3Q2YPC6 A0A1Y3EH44 A0A3Q3MD33 A0A3S1BQA2 A0A3B4WNY2 A0A2I4C6T6
PDB
2NSM
E-value=1.79938e-62,
Score=605
Ontologies
GO
PANTHER
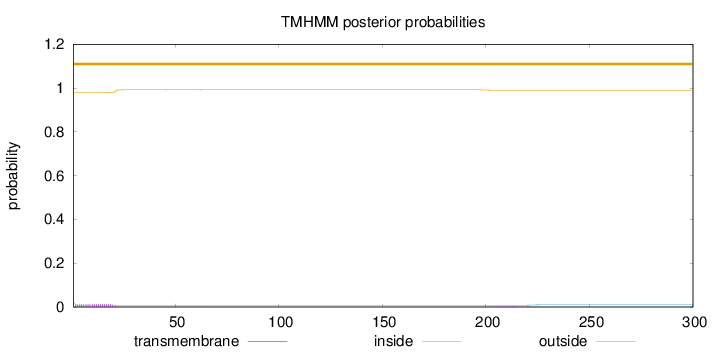
Topology
Subcellular location
Membrane
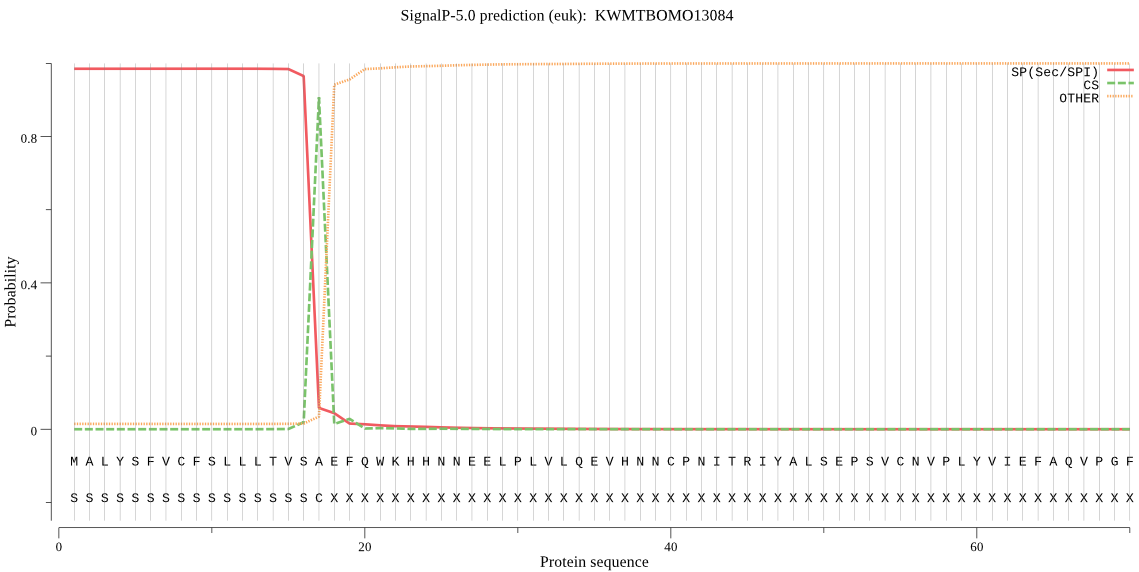
SignalP
Position: 1 - 17,
Likelihood: 0.985064
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.30997
Exp number, first 60 AAs:
0.25661
Total prob of N-in:
0.02113
outside
1 - 300
Population Genetic Test Statistics
Pi
15.653646
Theta
14.559839
Tajima's D
-0.425216
CLR
20.44172
CSRT
0.267786610669467
Interpretation
Uncertain
