Gene
KWMTBOMO13082
Pre Gene Modal
BGIBMGA000306
Annotation
PREDICTED:_uncharacterized_protein_LOC101740563_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.563
Sequence
CDS
ATGAAAATAAGCACGTTTATATATAAACCTTACGTATTATTGGATGTGGATACTGCAGTGGCACCGCTCGGAAGAGACGGTATAGAAATAAGAATGATTGACGAATTTTGCAGATGGATTAATTGTACTGTTCAAATTATAAGAGAAGATGTGGACTTATGGGGAGAAATTTATGAAAATGAAACAGGCATCGGGGTTATCGGAAGTGTTGTTGAAGGCCGGTCGGATATTGGAATTGCCGCTTTATATTCGTGGTATGAAGAATGGAAAGCCATGGATTTTTCAGTTTCTGTTGTCAGATCGGCTGTAATATGCTTGGTACCAGCTCCAAGAGTTTTGGAAAGTTGGGAGTTACCTTTTCTGCCATTCGGTAAAAGTATATGGATAGCTGTCGTGATTACATTTGTGTATGCCAGCATCGGTTTGACAATCGCTCAGGGATGTTCTTCTAACAAAGCTCTATTAATTGTCTTTGGTACTATAATTTCACAGTCTCAGTATATAGTAAGCGACTCTTGGCGCATTCGTTCTGTTATTGGTTGGCTTTTAGTTTCAAGTTTGATTTTGGTAAGTGCTTACGGAGCTGGACTTGCTTCTACATTTACTGTACCGCAGTATGAGCCATCTATTGACACTGTCCAAGATCTTCTCAACAGCCGCATGGAATGGGGTGCTAATCACGAAGCTTGGACATTTTCTCTAGCTTTATCTTCTGAGCCAGTAGCAAAGAAGTTAATTAAACAGTTTAAAATCTATTCTTTTGAAGAGTTACAGAGAAGAAGTTTCTTAAGGAAAATGGCCTTTAGCTTGGAAGAACTACCAGCAGGTATGTACGTGTGA
Protein
MKISTFIYKPYVLLDVDTAVAPLGRDGIEIRMIDEFCRWINCTVQIIREDVDLWGEIYENETGIGVIGSVVEGRSDIGIAALYSWYEEWKAMDFSVSVVRSAVICLVPAPRVLESWELPFLPFGKSIWIAVVITFVYASIGLTIAQGCSSNKALLIVFGTIISQSQYIVSDSWRIRSVIGWLLVSSLILVSAYGAGLASTFTVPQYEPSIDTVQDLLNSRMEWGANHEAWTFSLALSSEPVAKKLIKQFKIYSFEELQRRSFLRKMAFSLEELPAGMYV
Summary
Uniprot
H9ISS9
A0A2H1V256
A0A076E650
A0A2W1BJQ2
A0A2K8GL74
A0A194PWX0
+ More
A0A1B3B705 A0A194R146 E5FIA6 A0A0S1TPQ7 A0A386H9H8 A0A0K8TV01 A0A1B3P5G1 A0A1Y9TJK5 A0A146JVR8 A0A345BF28 A0A1Q1PP73 A0A0F7QEG2 A0A223HDA8 H9A5R8 A0A140G9G8 A0A076E637 A0A212F4A5 A0A223HCX0 A0A223HCX3 A0A0V0J120 A0A1Q1PP77 A0A3Q8HDQ2 W5J8R7 A0A2J7QRG6 A0A182SY81 A0A182FSF6 A0A182VRN4 B4J9E9 Q0C756 A0A182P639 A0A0U4VTB9 A0A182RUA9 A0A3B0J330 A0A182MC87 B4GBJ2 A0A182XV41 A0A182Q9U2 B5E0W1 A0A084VE37 A0A182U9I3 B4KLR5 A0A182HTW1 A0A182LG76 A0A0Q5WD24 A0A182VMS1 Q7PRA5 A0A182XHI5 A0A182JNI1 A0A182H8W6 B4MJ06 A0A182JE29 A0A182GHE2 A0A1W6L1B5 A0A0P8XRS8 B4LPH8 A0A182N544 B4IV73 A1Z6D6 A0A0M4ET77 A0A182GHE5 A0A1W7R6Y3 A0A182H8W4 Q0C751 A0A0K2D694 A0A067RCR0 A0A068FBN5 A0A0M5K6K1 A0A084WP99 Q0C757 U4UQR3 A0A1I8NEJ8 A0A1S4EUR5 A0A1I8P224 Q0C750 A0A2J7QQQ5 B4L0Y2 A0A2P8XLB8 A0A2P8Z0R0 A0A0M3QYQ6 A0A1S4EUU1 A0A1B0BE29 A0A3F2ZDJ5 A0A1I8NWW2 A0A2P8YUI3 A0A3F2ZDK2 R9PSR6 T1H854 T1H8Z0 B4LBN4 A0A3F2ZEC8 A0A2I4PHI1 B4IIQ3 A0A067R8D9 A0A182FQ68
A0A1B3B705 A0A194R146 E5FIA6 A0A0S1TPQ7 A0A386H9H8 A0A0K8TV01 A0A1B3P5G1 A0A1Y9TJK5 A0A146JVR8 A0A345BF28 A0A1Q1PP73 A0A0F7QEG2 A0A223HDA8 H9A5R8 A0A140G9G8 A0A076E637 A0A212F4A5 A0A223HCX0 A0A223HCX3 A0A0V0J120 A0A1Q1PP77 A0A3Q8HDQ2 W5J8R7 A0A2J7QRG6 A0A182SY81 A0A182FSF6 A0A182VRN4 B4J9E9 Q0C756 A0A182P639 A0A0U4VTB9 A0A182RUA9 A0A3B0J330 A0A182MC87 B4GBJ2 A0A182XV41 A0A182Q9U2 B5E0W1 A0A084VE37 A0A182U9I3 B4KLR5 A0A182HTW1 A0A182LG76 A0A0Q5WD24 A0A182VMS1 Q7PRA5 A0A182XHI5 A0A182JNI1 A0A182H8W6 B4MJ06 A0A182JE29 A0A182GHE2 A0A1W6L1B5 A0A0P8XRS8 B4LPH8 A0A182N544 B4IV73 A1Z6D6 A0A0M4ET77 A0A182GHE5 A0A1W7R6Y3 A0A182H8W4 Q0C751 A0A0K2D694 A0A067RCR0 A0A068FBN5 A0A0M5K6K1 A0A084WP99 Q0C757 U4UQR3 A0A1I8NEJ8 A0A1S4EUR5 A0A1I8P224 Q0C750 A0A2J7QQQ5 B4L0Y2 A0A2P8XLB8 A0A2P8Z0R0 A0A0M3QYQ6 A0A1S4EUU1 A0A1B0BE29 A0A3F2ZDJ5 A0A1I8NWW2 A0A2P8YUI3 A0A3F2ZDK2 R9PSR6 T1H854 T1H8Z0 B4LBN4 A0A3F2ZEC8 A0A2I4PHI1 B4IIQ3 A0A067R8D9 A0A182FQ68
Pubmed
19121390
24998398
28756777
29375398
26354079
21091811
+ More
26812239 26017144 27538507 29727827 28150741 25803580 22363688 27006164 27004525 22118469 20920257 23761445 17994087 17510324 26760975 25244985 15632085 24438588 20966253 12364791 14747013 17210077 26483478 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26265180 24845553 25880816 23537049 25315136 29403074 23517120
26812239 26017144 27538507 29727827 28150741 25803580 22363688 27006164 27004525 22118469 20920257 23761445 17994087 17510324 26760975 25244985 15632085 24438588 20966253 12364791 14747013 17210077 26483478 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26265180 24845553 25880816 23537049 25315136 29403074 23517120
EMBL
BABH01013177
ODYU01000339
SOQ34923.1
KF487728
AII01126.1
KZ150009
+ More
PZC75098.1 KY225531 ARO70543.1 KQ459586 KPI97856.1 KT588087 AOE47997.1 KQ460883 KPJ11427.1 HM562970 ADR64681.1 KR912015 ALM24942.1 MG546651 AYD42274.1 GCVX01000134 JAI18096.1 KX655897 AOG12846.1 KX084515 ARO76470.1 GEDO01000003 JAP88623.1 MG820681 AXF48852.1 KY325451 AQM73612.1 LC017786 BAR64800.1 KY283700 AST36359.1 JN836718 AFC91758.1 KU702620 AMM70647.1 KF487718 AII01116.1 AGBW02010414 OWR48561.1 KY283569 AST36229.1 KY283570 AST36230.1 GDKB01000020 JAP38476.1 KY325452 AQM73613.1 MF625609 AXY83436.1 ADMH02001856 ETN60852.1 NEVH01011896 PNF31178.1 CH916367 EDW01430.1 CH477186 EAT48971.1 FX982938 BAU20264.1 OUUW01000001 SPP75964.1 AXCM01004142 CH479181 EDW31287.1 AXCN02000174 AXCN02000175 CM000071 EDY68671.2 ATLV01012227 KE524773 KFB36231.1 CH933808 EDW09725.1 APCN01000586 CH954177 KQS71060.1 AAAB01008859 EAA07564.6 JXUM01029705 KQ560862 KXJ80588.1 CH963719 EDW72095.2 JXUM01063517 KQ562255 KXJ76291.1 KX609447 KB465964 ARN17854.1 RLZ02210.1 CH902619 KPU77322.1 CH940648 EDW61237.1 CH892654 EDX00750.2 AE013599 AAS64776.4 CP012524 ALC40675.1 JXUM01063522 KXJ76294.1 GEHC01000810 JAV46835.1 JXUM01029697 KXJ80586.1 EAT48976.1 KT279128 ALA15330.1 KK852549 KDR21537.1 KJ702122 AID61276.1 KP843217 ALD51353.1 ATLV01024999 KE525367 KFB52043.1 EAT48970.1 KB632326 ERL92496.1 EAT48977.1 NEVH01012083 PNF30918.1 CH933809 EDW19232.2 PYGN01001791 PSN32793.1 PYGN01000253 PSN50071.1 CP012526 ALC48029.1 JXJN01012751 JXJN01012752 AJWK01024855 PYGN01000353 PSN47912.1 GABX01000042 JAA74477.1 ACPB03022727 ACPB03026405 CH940647 EDW70844.2 AJVK01021584 KU523594 APZ81416.1 CH480844 EDW49824.1 KK852631 KDR19831.1
PZC75098.1 KY225531 ARO70543.1 KQ459586 KPI97856.1 KT588087 AOE47997.1 KQ460883 KPJ11427.1 HM562970 ADR64681.1 KR912015 ALM24942.1 MG546651 AYD42274.1 GCVX01000134 JAI18096.1 KX655897 AOG12846.1 KX084515 ARO76470.1 GEDO01000003 JAP88623.1 MG820681 AXF48852.1 KY325451 AQM73612.1 LC017786 BAR64800.1 KY283700 AST36359.1 JN836718 AFC91758.1 KU702620 AMM70647.1 KF487718 AII01116.1 AGBW02010414 OWR48561.1 KY283569 AST36229.1 KY283570 AST36230.1 GDKB01000020 JAP38476.1 KY325452 AQM73613.1 MF625609 AXY83436.1 ADMH02001856 ETN60852.1 NEVH01011896 PNF31178.1 CH916367 EDW01430.1 CH477186 EAT48971.1 FX982938 BAU20264.1 OUUW01000001 SPP75964.1 AXCM01004142 CH479181 EDW31287.1 AXCN02000174 AXCN02000175 CM000071 EDY68671.2 ATLV01012227 KE524773 KFB36231.1 CH933808 EDW09725.1 APCN01000586 CH954177 KQS71060.1 AAAB01008859 EAA07564.6 JXUM01029705 KQ560862 KXJ80588.1 CH963719 EDW72095.2 JXUM01063517 KQ562255 KXJ76291.1 KX609447 KB465964 ARN17854.1 RLZ02210.1 CH902619 KPU77322.1 CH940648 EDW61237.1 CH892654 EDX00750.2 AE013599 AAS64776.4 CP012524 ALC40675.1 JXUM01063522 KXJ76294.1 GEHC01000810 JAV46835.1 JXUM01029697 KXJ80586.1 EAT48976.1 KT279128 ALA15330.1 KK852549 KDR21537.1 KJ702122 AID61276.1 KP843217 ALD51353.1 ATLV01024999 KE525367 KFB52043.1 EAT48970.1 KB632326 ERL92496.1 EAT48977.1 NEVH01012083 PNF30918.1 CH933809 EDW19232.2 PYGN01001791 PSN32793.1 PYGN01000253 PSN50071.1 CP012526 ALC48029.1 JXJN01012751 JXJN01012752 AJWK01024855 PYGN01000353 PSN47912.1 GABX01000042 JAA74477.1 ACPB03022727 ACPB03026405 CH940647 EDW70844.2 AJVK01021584 KU523594 APZ81416.1 CH480844 EDW49824.1 KK852631 KDR19831.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000000673
UP000235965
+ More
UP000075901 UP000069272 UP000075920 UP000001070 UP000008820 UP000075885 UP000075900 UP000268350 UP000075883 UP000008744 UP000076408 UP000075886 UP000001819 UP000030765 UP000075902 UP000009192 UP000075840 UP000075882 UP000008711 UP000075903 UP000007062 UP000076407 UP000075881 UP000069940 UP000249989 UP000007798 UP000075880 UP000007801 UP000008792 UP000075884 UP000002282 UP000000803 UP000092553 UP000027135 UP000030742 UP000095301 UP000095300 UP000245037 UP000092460 UP000092461 UP000015103 UP000092462 UP000001292
UP000075901 UP000069272 UP000075920 UP000001070 UP000008820 UP000075885 UP000075900 UP000268350 UP000075883 UP000008744 UP000076408 UP000075886 UP000001819 UP000030765 UP000075902 UP000009192 UP000075840 UP000075882 UP000008711 UP000075903 UP000007062 UP000076407 UP000075881 UP000069940 UP000249989 UP000007798 UP000075880 UP000007801 UP000008792 UP000075884 UP000002282 UP000000803 UP000092553 UP000027135 UP000030742 UP000095301 UP000095300 UP000245037 UP000092460 UP000092461 UP000015103 UP000092462 UP000001292
Interpro
SUPFAM
SSF49464
SSF49464
ProteinModelPortal
H9ISS9
A0A2H1V256
A0A076E650
A0A2W1BJQ2
A0A2K8GL74
A0A194PWX0
+ More
A0A1B3B705 A0A194R146 E5FIA6 A0A0S1TPQ7 A0A386H9H8 A0A0K8TV01 A0A1B3P5G1 A0A1Y9TJK5 A0A146JVR8 A0A345BF28 A0A1Q1PP73 A0A0F7QEG2 A0A223HDA8 H9A5R8 A0A140G9G8 A0A076E637 A0A212F4A5 A0A223HCX0 A0A223HCX3 A0A0V0J120 A0A1Q1PP77 A0A3Q8HDQ2 W5J8R7 A0A2J7QRG6 A0A182SY81 A0A182FSF6 A0A182VRN4 B4J9E9 Q0C756 A0A182P639 A0A0U4VTB9 A0A182RUA9 A0A3B0J330 A0A182MC87 B4GBJ2 A0A182XV41 A0A182Q9U2 B5E0W1 A0A084VE37 A0A182U9I3 B4KLR5 A0A182HTW1 A0A182LG76 A0A0Q5WD24 A0A182VMS1 Q7PRA5 A0A182XHI5 A0A182JNI1 A0A182H8W6 B4MJ06 A0A182JE29 A0A182GHE2 A0A1W6L1B5 A0A0P8XRS8 B4LPH8 A0A182N544 B4IV73 A1Z6D6 A0A0M4ET77 A0A182GHE5 A0A1W7R6Y3 A0A182H8W4 Q0C751 A0A0K2D694 A0A067RCR0 A0A068FBN5 A0A0M5K6K1 A0A084WP99 Q0C757 U4UQR3 A0A1I8NEJ8 A0A1S4EUR5 A0A1I8P224 Q0C750 A0A2J7QQQ5 B4L0Y2 A0A2P8XLB8 A0A2P8Z0R0 A0A0M3QYQ6 A0A1S4EUU1 A0A1B0BE29 A0A3F2ZDJ5 A0A1I8NWW2 A0A2P8YUI3 A0A3F2ZDK2 R9PSR6 T1H854 T1H8Z0 B4LBN4 A0A3F2ZEC8 A0A2I4PHI1 B4IIQ3 A0A067R8D9 A0A182FQ68
A0A1B3B705 A0A194R146 E5FIA6 A0A0S1TPQ7 A0A386H9H8 A0A0K8TV01 A0A1B3P5G1 A0A1Y9TJK5 A0A146JVR8 A0A345BF28 A0A1Q1PP73 A0A0F7QEG2 A0A223HDA8 H9A5R8 A0A140G9G8 A0A076E637 A0A212F4A5 A0A223HCX0 A0A223HCX3 A0A0V0J120 A0A1Q1PP77 A0A3Q8HDQ2 W5J8R7 A0A2J7QRG6 A0A182SY81 A0A182FSF6 A0A182VRN4 B4J9E9 Q0C756 A0A182P639 A0A0U4VTB9 A0A182RUA9 A0A3B0J330 A0A182MC87 B4GBJ2 A0A182XV41 A0A182Q9U2 B5E0W1 A0A084VE37 A0A182U9I3 B4KLR5 A0A182HTW1 A0A182LG76 A0A0Q5WD24 A0A182VMS1 Q7PRA5 A0A182XHI5 A0A182JNI1 A0A182H8W6 B4MJ06 A0A182JE29 A0A182GHE2 A0A1W6L1B5 A0A0P8XRS8 B4LPH8 A0A182N544 B4IV73 A1Z6D6 A0A0M4ET77 A0A182GHE5 A0A1W7R6Y3 A0A182H8W4 Q0C751 A0A0K2D694 A0A067RCR0 A0A068FBN5 A0A0M5K6K1 A0A084WP99 Q0C757 U4UQR3 A0A1I8NEJ8 A0A1S4EUR5 A0A1I8P224 Q0C750 A0A2J7QQQ5 B4L0Y2 A0A2P8XLB8 A0A2P8Z0R0 A0A0M3QYQ6 A0A1S4EUU1 A0A1B0BE29 A0A3F2ZDJ5 A0A1I8NWW2 A0A2P8YUI3 A0A3F2ZDK2 R9PSR6 T1H854 T1H8Z0 B4LBN4 A0A3F2ZEC8 A0A2I4PHI1 B4IIQ3 A0A067R8D9 A0A182FQ68
PDB
5KUF
E-value=2.50165e-09,
Score=147
Ontologies
GO
PANTHER
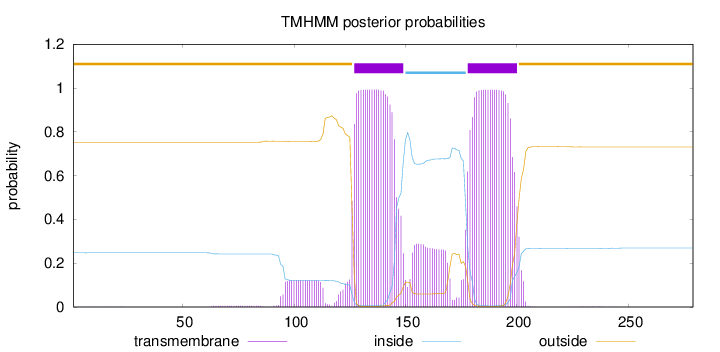
Topology
Length:
279
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
50.4214200000001
Exp number, first 60 AAs:
0.01107
Total prob of N-in:
0.24773
outside
1 - 126
TMhelix
127 - 149
inside
150 - 177
TMhelix
178 - 200
outside
201 - 279
Population Genetic Test Statistics
Pi
184.155627
Theta
174.691524
Tajima's D
0.544394
CLR
0
CSRT
0.526773661316934
Interpretation
Uncertain
