Gene
KWMTBOMO13079
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_integrator_complex_subunit_12_[Bombyx_mori]
Full name
Integrator complex subunit 12
Location in the cell
Nuclear Reliability : 4.145
Sequence
CDS
ATGTCATCAATTGATTTCGATCCAGCTATAAAATTATGTTTAAAATACTTACATTCAAGTTCTGTAGATTCTACGGAACAATTGCGATTGACTCTCGATGATCACATCAGACAATCATATGGTAGTGCAAAAACTCTTGGTAATATGTTACCGAAGAAATATCTTAACGAAGAGAAGATGGAGTCACCTAGATCCAAACACAAAATAGATAAGAATGCAGTAACTAAGACGGTTCCTGTACCACAACAAAGTCCACAACAAATTCAAATACCTGAGAGAGAAAACGATGATGGTTCAGTTATGGATGGAGAATTGGCTTTTGACTTGTTAGAGGAAGACTTGACTTGTGCTGTCTGTAGACAAATAGCTGTGCAAGCCGGAAATCGCTTAGTGGAGTGTGATGGGTGCCGGGCACTGTACCATCAGGATTGTCATAAGCCTGTGATCAGTGACAATGATATCGCTGCAGGTTGGCAATGTGCAACCTGTCTTGCAGCACATGGATTTACAACTAATTATTCAAGTAAAACTTCATCGAGCTCCAAGTCACCATCAAGAGTGTCTGGTTCCACAACCCCTGTAAAAATTTCATCTAGCAGTTCATCTTCATCATCACCATCGAAAGTTGTTACTCCAAATATGAATATCATTTCTGCAGACAAAAGGTTGCAGATAATGAAGAAGAAAGCTGCAAAACAACATGAAAAGAAGAAAAGCTCTAAGTCGTAA
Protein
MSSIDFDPAIKLCLKYLHSSSVDSTEQLRLTLDDHIRQSYGSAKTLGNMLPKKYLNEEKMESPRSKHKIDKNAVTKTVPVPQQSPQQIQIPERENDDGSVMDGELAFDLLEEDLTCAVCRQIAVQAGNRLVECDGCRALYHQDCHKPVISDNDIAAGWQCATCLAAHGFTTNYSSKTSSSSKSPSRVSGSTTPVKISSSSSSSSSPSKVVTPNMNIISADKRLQIMKKKAAKQHEKKKSSKS
Summary
Description
Component of the Integrator complex, a complex involved in the transcription of small nuclear RNAs (snRNA) and their 3'-box-dependent processing (PubMed:21078872, PubMed:23097424, PubMed:23288851). Involved in the 3'-end processing of the U7 snRNA, and also the spliceosomal snRNAs U1, U2, U4 and U5 (PubMed:21078872, PubMed:23097424, PubMed:23288851). Required for the normal expression of the Integrator complex component IntS1 (PubMed:23288851). May mediate recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (By similarity).
Subunit
Belongs to the multiprotein complex Integrator, at least composed of IntS1, IntS2, IntS3, IntS4, omd/IntS5, IntS6, defl/IntS7, IntS8, IntS9, IntS10, IntS11, IntS12, asun/IntS13 and IntS14 (PubMed:23097424). Within the complex, interacts with IntS1 and IntS9 (PubMed:23288851). Interaction with IntS1 is likely to be important for promoting 3'-end processing of snRNAs (PubMed:23288851).
Similarity
Belongs to the Integrator subunit 12 family.
Keywords
Complete proteome
Metal-binding
Nucleus
Reference proteome
Zinc
Zinc-finger
Feature
chain Integrator complex subunit 12
Uniprot
A0A1E1WKX3
S4PHC4
A0A3S2TAL7
A0A2H1V257
A0A2W1BP86
A0A2A4J0L5
+ More
A0A194PXV1 A0A194R2F5 H9ISS7 A0A3L8DX39 A0A026WDK3 A0A310SSQ7 A0A182R8H4 A0A084WQ69 A0A182MWM9 A0A182W341 E2ADR6 A0A0J7KNG3 A0A0M8ZRP6 A0A182PG82 A0A0L7R303 A0A182HVJ6 Q7QA92 A0A182UWA0 A0A1Q3F1Y1 A0A182XF28 A0A182LNJ5 A0A182TWX5 A0A088A4N1 A0A1B0D4L3 A0A2A3E9B1 A0A195B0W7 A0A151IEY6 F4WPF2 A0A158NR00 A0A195E6I2 B0WMX5 A0A195EUD0 A0A151WMC7 Q170V4 A0A182YJQ0 E9ISL8 A0A182K051 A0A182QGN7 A0A023EP97 A0A182G6H7 E2BN22 A0A182NCQ6 A0A232EJR5 A0A182IPC5 A0A0P6IU04 A0A1S4FHT6 A0A2M4APF1 A0A1B0CAG0 W5JET0 A0A182FPJ7 A0A1L8DCI5 Q9VBB3 A0A0M4EIX1 A0A0L0CQ62 B3P8A1 B4QWS4 B4PSC5 B4IBZ7 A0A1W4VKF2 W8C9L1 B3M3B9 E0VMJ4 A0A0K8ULE7 A0A034WKR7 T1JLY1 B4JY74 G1QCI4 G9K5W7 G5AT22 A0A3Q1HSR5 S9YB32 G1PIZ5 A0A3Q7P609 A0A340WYU8 A0A1L8HV76 A0A2Y9N0D8 A0A341C618 K9J136 A0A2U4AQT2 A0A1U7SW01 A0A2U4AR75
A0A194PXV1 A0A194R2F5 H9ISS7 A0A3L8DX39 A0A026WDK3 A0A310SSQ7 A0A182R8H4 A0A084WQ69 A0A182MWM9 A0A182W341 E2ADR6 A0A0J7KNG3 A0A0M8ZRP6 A0A182PG82 A0A0L7R303 A0A182HVJ6 Q7QA92 A0A182UWA0 A0A1Q3F1Y1 A0A182XF28 A0A182LNJ5 A0A182TWX5 A0A088A4N1 A0A1B0D4L3 A0A2A3E9B1 A0A195B0W7 A0A151IEY6 F4WPF2 A0A158NR00 A0A195E6I2 B0WMX5 A0A195EUD0 A0A151WMC7 Q170V4 A0A182YJQ0 E9ISL8 A0A182K051 A0A182QGN7 A0A023EP97 A0A182G6H7 E2BN22 A0A182NCQ6 A0A232EJR5 A0A182IPC5 A0A0P6IU04 A0A1S4FHT6 A0A2M4APF1 A0A1B0CAG0 W5JET0 A0A182FPJ7 A0A1L8DCI5 Q9VBB3 A0A0M4EIX1 A0A0L0CQ62 B3P8A1 B4QWS4 B4PSC5 B4IBZ7 A0A1W4VKF2 W8C9L1 B3M3B9 E0VMJ4 A0A0K8ULE7 A0A034WKR7 T1JLY1 B4JY74 G1QCI4 G9K5W7 G5AT22 A0A3Q1HSR5 S9YB32 G1PIZ5 A0A3Q7P609 A0A340WYU8 A0A1L8HV76 A0A2Y9N0D8 A0A341C618 K9J136 A0A2U4AQT2 A0A1U7SW01 A0A2U4AR75
Pubmed
23622113
28756777
26354079
19121390
30249741
24508170
+ More
24438588 20798317 12364791 14747013 17210077 20966253 21719571 21347285 17510324 25244985 21282665 24945155 26483478 28648823 26999592 20920257 23761445 10731132 12537572 12537569 21078872 23097424 23288851 26108605 17994087 17550304 24495485 20566863 25348373 21993624 23236062 21993625 23149746 27762356
24438588 20798317 12364791 14747013 17210077 20966253 21719571 21347285 17510324 25244985 21282665 24945155 26483478 28648823 26999592 20920257 23761445 10731132 12537572 12537569 21078872 23097424 23288851 26108605 17994087 17550304 24495485 20566863 25348373 21993624 23236062 21993625 23149746 27762356
EMBL
GDQN01003400
JAT87654.1
GAIX01003327
JAA89233.1
RSAL01002880
RVE40395.1
+ More
ODYU01000339 SOQ34920.1 KZ150009 PZC75117.1 NWSH01004475 PCG65064.1 KQ459586 KPI97853.1 KQ460883 KPJ11430.1 BABH01013178 QOIP01000003 RLU25011.1 KK107273 EZA53761.1 KQ760801 OAD58966.1 ATLV01025277 KE525379 KFB52363.1 AXCM01000759 GL438820 EFN68428.1 LBMM01005008 KMQ91918.1 KQ435885 KOX69587.1 KQ414663 KOC65260.1 APCN01005788 AAAB01008898 EAA09080.4 GFDL01013480 JAV21565.1 AJVK01024655 KZ288315 PBC28315.1 KQ976691 KYM77937.1 KQ977835 KYM99375.1 GL888243 EGI64047.1 ADTU01023715 KQ979608 KYN20454.1 DS232003 EDS31367.1 KQ981965 KYN31860.1 KQ982944 KYQ48993.1 CH477466 EAT40483.1 GL765434 EFZ16312.1 AXCN02000334 GAPW01002301 JAC11297.1 JXUM01045220 KQ561430 KXJ78610.1 GL449382 EFN82926.1 NNAY01003972 OXU18551.1 GDUN01000704 JAN95215.1 GGFK01009343 MBW42664.1 AJWK01003749 ADMH02001712 ETN61329.1 GFDF01009922 JAV04162.1 AE014297 AY070574 AAL48045.1 CP012526 ALC46721.1 JRES01000062 KNC34505.1 CH954182 EDV53505.1 CM000364 EDX14591.1 CM000160 EDW98587.1 CH480828 EDW45155.1 GAMC01002064 JAC04492.1 CH902617 EDV43580.1 DS235313 EEB14600.1 GDHF01024996 JAI27318.1 GAKP01004576 GAKP01004574 JAC54378.1 JH431482 CH916377 EDV90636.1 AAPE02006782 JP011694 AES00292.1 JH166824 GEBF01007386 EHB00183.1 JAN96246.1 KB017014 EPY81200.1 AAPE02032741 CM004466 OCT99918.1 GABZ01005966 JAA47559.1
ODYU01000339 SOQ34920.1 KZ150009 PZC75117.1 NWSH01004475 PCG65064.1 KQ459586 KPI97853.1 KQ460883 KPJ11430.1 BABH01013178 QOIP01000003 RLU25011.1 KK107273 EZA53761.1 KQ760801 OAD58966.1 ATLV01025277 KE525379 KFB52363.1 AXCM01000759 GL438820 EFN68428.1 LBMM01005008 KMQ91918.1 KQ435885 KOX69587.1 KQ414663 KOC65260.1 APCN01005788 AAAB01008898 EAA09080.4 GFDL01013480 JAV21565.1 AJVK01024655 KZ288315 PBC28315.1 KQ976691 KYM77937.1 KQ977835 KYM99375.1 GL888243 EGI64047.1 ADTU01023715 KQ979608 KYN20454.1 DS232003 EDS31367.1 KQ981965 KYN31860.1 KQ982944 KYQ48993.1 CH477466 EAT40483.1 GL765434 EFZ16312.1 AXCN02000334 GAPW01002301 JAC11297.1 JXUM01045220 KQ561430 KXJ78610.1 GL449382 EFN82926.1 NNAY01003972 OXU18551.1 GDUN01000704 JAN95215.1 GGFK01009343 MBW42664.1 AJWK01003749 ADMH02001712 ETN61329.1 GFDF01009922 JAV04162.1 AE014297 AY070574 AAL48045.1 CP012526 ALC46721.1 JRES01000062 KNC34505.1 CH954182 EDV53505.1 CM000364 EDX14591.1 CM000160 EDW98587.1 CH480828 EDW45155.1 GAMC01002064 JAC04492.1 CH902617 EDV43580.1 DS235313 EEB14600.1 GDHF01024996 JAI27318.1 GAKP01004576 GAKP01004574 JAC54378.1 JH431482 CH916377 EDV90636.1 AAPE02006782 JP011694 AES00292.1 JH166824 GEBF01007386 EHB00183.1 JAN96246.1 KB017014 EPY81200.1 AAPE02032741 CM004466 OCT99918.1 GABZ01005966 JAA47559.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000005204
UP000279307
+ More
UP000053097 UP000075900 UP000030765 UP000075883 UP000075920 UP000000311 UP000036403 UP000053105 UP000075885 UP000053825 UP000075840 UP000007062 UP000075903 UP000076407 UP000075882 UP000075902 UP000005203 UP000092462 UP000242457 UP000078540 UP000078542 UP000007755 UP000005205 UP000078492 UP000002320 UP000078541 UP000075809 UP000008820 UP000076408 UP000075881 UP000075886 UP000069940 UP000249989 UP000008237 UP000075884 UP000215335 UP000075880 UP000092461 UP000000673 UP000069272 UP000000803 UP000092553 UP000037069 UP000008711 UP000000304 UP000002282 UP000001292 UP000192221 UP000007801 UP000009046 UP000001070 UP000001074 UP000006813 UP000257200 UP000286641 UP000265300 UP000186698 UP000248483 UP000252040 UP000245320 UP000189704
UP000053097 UP000075900 UP000030765 UP000075883 UP000075920 UP000000311 UP000036403 UP000053105 UP000075885 UP000053825 UP000075840 UP000007062 UP000075903 UP000076407 UP000075882 UP000075902 UP000005203 UP000092462 UP000242457 UP000078540 UP000078542 UP000007755 UP000005205 UP000078492 UP000002320 UP000078541 UP000075809 UP000008820 UP000076408 UP000075881 UP000075886 UP000069940 UP000249989 UP000008237 UP000075884 UP000215335 UP000075880 UP000092461 UP000000673 UP000069272 UP000000803 UP000092553 UP000037069 UP000008711 UP000000304 UP000002282 UP000001292 UP000192221 UP000007801 UP000009046 UP000001070 UP000001074 UP000006813 UP000257200 UP000286641 UP000265300 UP000186698 UP000248483 UP000252040 UP000245320 UP000189704
Interpro
IPR019787
Znf_PHD-finger
+ More
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR039053 Int12
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR039054 Int12_PHD
IPR006094 Oxid_FAD_bind_N
IPR036318 FAD-bd_PCMH-like_sf
IPR016166 FAD-bd_PCMH
IPR025650 Alkyl-DHAP_Synthase
IPR016171 Vanillyl_alc_oxidase_C-sub2
IPR016164 FAD-linked_Oxase-like_C
IPR004113 FAD-linked_oxidase_C
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR009011 Man6P_isomerase_rcpt-bd_dom_sf
IPR000479 CIMR
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR039053 Int12
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR039054 Int12_PHD
IPR006094 Oxid_FAD_bind_N
IPR036318 FAD-bd_PCMH-like_sf
IPR016166 FAD-bd_PCMH
IPR025650 Alkyl-DHAP_Synthase
IPR016171 Vanillyl_alc_oxidase_C-sub2
IPR016164 FAD-linked_Oxase-like_C
IPR004113 FAD-linked_oxidase_C
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR009011 Man6P_isomerase_rcpt-bd_dom_sf
IPR000479 CIMR
ProteinModelPortal
A0A1E1WKX3
S4PHC4
A0A3S2TAL7
A0A2H1V257
A0A2W1BP86
A0A2A4J0L5
+ More
A0A194PXV1 A0A194R2F5 H9ISS7 A0A3L8DX39 A0A026WDK3 A0A310SSQ7 A0A182R8H4 A0A084WQ69 A0A182MWM9 A0A182W341 E2ADR6 A0A0J7KNG3 A0A0M8ZRP6 A0A182PG82 A0A0L7R303 A0A182HVJ6 Q7QA92 A0A182UWA0 A0A1Q3F1Y1 A0A182XF28 A0A182LNJ5 A0A182TWX5 A0A088A4N1 A0A1B0D4L3 A0A2A3E9B1 A0A195B0W7 A0A151IEY6 F4WPF2 A0A158NR00 A0A195E6I2 B0WMX5 A0A195EUD0 A0A151WMC7 Q170V4 A0A182YJQ0 E9ISL8 A0A182K051 A0A182QGN7 A0A023EP97 A0A182G6H7 E2BN22 A0A182NCQ6 A0A232EJR5 A0A182IPC5 A0A0P6IU04 A0A1S4FHT6 A0A2M4APF1 A0A1B0CAG0 W5JET0 A0A182FPJ7 A0A1L8DCI5 Q9VBB3 A0A0M4EIX1 A0A0L0CQ62 B3P8A1 B4QWS4 B4PSC5 B4IBZ7 A0A1W4VKF2 W8C9L1 B3M3B9 E0VMJ4 A0A0K8ULE7 A0A034WKR7 T1JLY1 B4JY74 G1QCI4 G9K5W7 G5AT22 A0A3Q1HSR5 S9YB32 G1PIZ5 A0A3Q7P609 A0A340WYU8 A0A1L8HV76 A0A2Y9N0D8 A0A341C618 K9J136 A0A2U4AQT2 A0A1U7SW01 A0A2U4AR75
A0A194PXV1 A0A194R2F5 H9ISS7 A0A3L8DX39 A0A026WDK3 A0A310SSQ7 A0A182R8H4 A0A084WQ69 A0A182MWM9 A0A182W341 E2ADR6 A0A0J7KNG3 A0A0M8ZRP6 A0A182PG82 A0A0L7R303 A0A182HVJ6 Q7QA92 A0A182UWA0 A0A1Q3F1Y1 A0A182XF28 A0A182LNJ5 A0A182TWX5 A0A088A4N1 A0A1B0D4L3 A0A2A3E9B1 A0A195B0W7 A0A151IEY6 F4WPF2 A0A158NR00 A0A195E6I2 B0WMX5 A0A195EUD0 A0A151WMC7 Q170V4 A0A182YJQ0 E9ISL8 A0A182K051 A0A182QGN7 A0A023EP97 A0A182G6H7 E2BN22 A0A182NCQ6 A0A232EJR5 A0A182IPC5 A0A0P6IU04 A0A1S4FHT6 A0A2M4APF1 A0A1B0CAG0 W5JET0 A0A182FPJ7 A0A1L8DCI5 Q9VBB3 A0A0M4EIX1 A0A0L0CQ62 B3P8A1 B4QWS4 B4PSC5 B4IBZ7 A0A1W4VKF2 W8C9L1 B3M3B9 E0VMJ4 A0A0K8ULE7 A0A034WKR7 T1JLY1 B4JY74 G1QCI4 G9K5W7 G5AT22 A0A3Q1HSR5 S9YB32 G1PIZ5 A0A3Q7P609 A0A340WYU8 A0A1L8HV76 A0A2Y9N0D8 A0A341C618 K9J136 A0A2U4AQT2 A0A1U7SW01 A0A2U4AR75
PDB
1WEV
E-value=4.27836e-11,
Score=161
Ontologies
GO
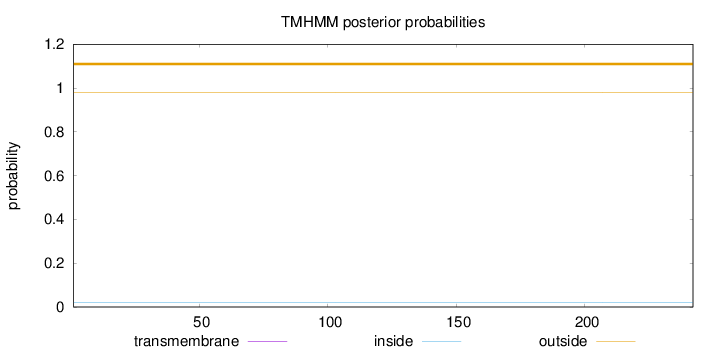
Topology
Subcellular location
Nucleus
Length:
242
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00197
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02138
outside
1 - 242
Population Genetic Test Statistics
Pi
314.632242
Theta
209.801515
Tajima's D
1.834132
CLR
0
CSRT
0.855757212139393
Interpretation
Uncertain
