Gene
KWMTBOMO13077
Pre Gene Modal
BGIBMGA000303
Annotation
PREDICTED:_GTP-binding_protein_Di-Ras2_[Amyelois_transitella]
Full name
GTP-binding protein Di-Ras2
Alternative Name
Distinct subgroup of the Ras family member 2
Location in the cell
Nuclear Reliability : 2.145
Sequence
CDS
ATGCCGGAACAAAGCAATGATTATCGCGTCGTAGTTTTTGGTGCGGGAGGTGTTGGTAAAAGTTCTCTAGTTCTCCGTTTCGTTAAAGGAACATTTAGAGAGTCTTACATACCAACAATAGAAGACACATATAGACAGGTGATAAGCAGTAACAAAACAATATGCACATTACAAATTACTGATACTACTGGTTCACATCAGTTTCCGGCTATGCAAAGACTGTCTATAAGCAAGGGCCACGCTTTTATATTGGTTTACTCCGTGAGCAGTCGACAGTCACTAGAAGAATTGAAACCAATATGGCAAACAATCAAAGAAATAAAAGGCGTGGAGCTCCCTAACATCCCTGTGATGTTGGCAGGCAACAAATGTGATGAGAGCCCAGAGATTCGAGAAGTATCAGCTTCTGAAGGACAGGCGCAGGCGCAAGCCTGGGGCGTATCCTTCATGGAGACATCTGCTAAAACGAATCACAATGTAACACAACTCTTTCAAGAGCTATTGAACATGGAAAAGAACAGAAACGTGTCTCTTCAAGTTGATGGAAAAGGTAAGAAGAAGAAAGATAAAGTTGTGAAAGAGACAGGAGAAGCTTCTGCTGCCGGCAAAGAGAAATGTCTTGTTATGTAA
Protein
MPEQSNDYRVVVFGAGGVGKSSLVLRFVKGTFRESYIPTIEDTYRQVISSNKTICTLQITDTTGSHQFPAMQRLSISKGHAFILVYSVSSRQSLEELKPIWQTIKEIKGVELPNIPVMLAGNKCDESPEIREVSASEGQAQAQAWGVSFMETSAKTNHNVTQLFQELLNMEKNRNVSLQVDGKGKKKKDKVVKETGEASAAGKEKCLVM
Summary
Description
Displays low GTPase activity and exists predominantly in the GTP-bound form.
Similarity
Belongs to the small GTPase superfamily. Di-Ras family.
Keywords
Cell membrane
Complete proteome
GTP-binding
Lipoprotein
Membrane
Methylation
Nucleotide-binding
Phosphoprotein
Prenylation
3D-structure
Reference proteome
Feature
chain GTP-binding protein Di-Ras2
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
H9ISS6
A0A2W1BJA6
A0A2H1V241
A0A194PWW5
A0A194R0W0
A0A212F489
+ More
A0A0L7L0A3 A0A1W4WM32 A0A1J1J484 D6X324 B3M1C7 Q299I2 A0A1Y1JQS7 A0A1W4W0V2 Q9VH66 B4PVF8 B3P4Q2 W8BLM8 A0A3B0KRD2 B4N810 A0A034V216 B4LYX2 A0A1U7UQZ4 A0A286XUM9 Q95KD9 U4UJK8 A0A091DHC1 N6TUB6 A0A2Y9E698 L8YAR4 G3HIA6 A0A2K6AND2 A0A2K6JVH6 A0A2I2Y9Y4 A0A2I3HQU6 A0A2K5XXR9 A0A0D9SB09 A0A2R8ZHX2 A0A096NE98 A0A2K5DZN0 A0A2K6NIW5 A0A2K5L062 A0A2K5H7K0 A0A2K5PA64 A0A2K6S7X6 H2PSM8 G7PSP0 F7EK27 A0A2I3SKK7 F7FTA1 Q96HU8 G5C2H5 A0A1S3G0J3 G1SCX2 E0VEI8 D3ZHX3 Q3UWU7 A0A1U8BF78 Q5PR73 F7DZR6 A0A287AKH0 A0A340WB13 A0A2Y9FDB2 A0A2F0AXQ5 A0A384B2G5 G1PCU9 S7NNM2 G3TH62 B4JV68 I3MX94 Q5R6S2 A0A1S3AT03 A0A341ACT5 A0A2U4BF66 A0A2Y9M5Q2 A0A2U3XPN3 A0A2Y9K525 A0A2U3WLR0 M3Z710 S9XB92 A0A2Y9HE79 A0A3Q7QMT5 H0XIA3 A0A2K6GK24 A0A384C2L9 E2RK40 A0A3Q7RCR3 A0A3Q7Y3A8 D2HB69 L5MK87 M3WE12 A0A224XX81 A0A182T4X9 Q007S9 A0A182WS04 A0A182UYI2 A0A182KVY0 A0A182TWS6 A0A182ICW6 Q7QI45
A0A0L7L0A3 A0A1W4WM32 A0A1J1J484 D6X324 B3M1C7 Q299I2 A0A1Y1JQS7 A0A1W4W0V2 Q9VH66 B4PVF8 B3P4Q2 W8BLM8 A0A3B0KRD2 B4N810 A0A034V216 B4LYX2 A0A1U7UQZ4 A0A286XUM9 Q95KD9 U4UJK8 A0A091DHC1 N6TUB6 A0A2Y9E698 L8YAR4 G3HIA6 A0A2K6AND2 A0A2K6JVH6 A0A2I2Y9Y4 A0A2I3HQU6 A0A2K5XXR9 A0A0D9SB09 A0A2R8ZHX2 A0A096NE98 A0A2K5DZN0 A0A2K6NIW5 A0A2K5L062 A0A2K5H7K0 A0A2K5PA64 A0A2K6S7X6 H2PSM8 G7PSP0 F7EK27 A0A2I3SKK7 F7FTA1 Q96HU8 G5C2H5 A0A1S3G0J3 G1SCX2 E0VEI8 D3ZHX3 Q3UWU7 A0A1U8BF78 Q5PR73 F7DZR6 A0A287AKH0 A0A340WB13 A0A2Y9FDB2 A0A2F0AXQ5 A0A384B2G5 G1PCU9 S7NNM2 G3TH62 B4JV68 I3MX94 Q5R6S2 A0A1S3AT03 A0A341ACT5 A0A2U4BF66 A0A2Y9M5Q2 A0A2U3XPN3 A0A2Y9K525 A0A2U3WLR0 M3Z710 S9XB92 A0A2Y9HE79 A0A3Q7QMT5 H0XIA3 A0A2K6GK24 A0A384C2L9 E2RK40 A0A3Q7RCR3 A0A3Q7Y3A8 D2HB69 L5MK87 M3WE12 A0A224XX81 A0A182T4X9 Q007S9 A0A182WS04 A0A182UYI2 A0A182KVY0 A0A182TWS6 A0A182ICW6 Q7QI45
Pubmed
19121390
28756777
26354079
22118469
26227816
18362917
+ More
19820115 17994087 15632085 23185243 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 25348373 21993624 23537049 23385571 21804562 22398555 22722832 25362486 22002653 17431167 25319552 16136131 25243066 12194967 14702039 15164053 15489334 21993625 20566863 15057822 10349636 11042159 11076861 11217851 12466851 16141073 21183079 19892987 30723633 23149746 24813606 16341006 20010809 17975172 20966253 12364791 14747013 17210077
19820115 17994087 15632085 23185243 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 25348373 21993624 23537049 23385571 21804562 22398555 22722832 25362486 22002653 17431167 25319552 16136131 25243066 12194967 14702039 15164053 15489334 21993625 20566863 15057822 10349636 11042159 11076861 11217851 12466851 16141073 21183079 19892987 30723633 23149746 24813606 16341006 20010809 17975172 20966253 12364791 14747013 17210077
EMBL
BABH01013178
KZ150009
PZC75119.1
ODYU01000339
SOQ34918.1
KQ459586
+ More
KPI97851.1 KQ460883 KPJ11433.1 AGBW02010414 OWR48556.1 JTDY01003999 KOB68731.1 CVRI01000070 CRL07207.1 KQ971372 EFA10311.1 CH902617 EDV42154.1 CM000070 EAL27721.2 KRT00249.1 KRT00250.1 GEZM01103095 GEZM01103093 JAV51564.1 AE014297 BT149876 AAF54453.1 AGC12536.1 ALI30550.1 CM000160 EDW96731.1 CH954181 EDV49705.1 GAMC01008633 GAMC01008632 GAMC01008631 GAMC01008630 JAB97923.1 OUUW01000013 SPP87801.1 CH964232 EDW81261.1 GAKP01022780 JAC36172.1 CH940650 EDW68075.1 AAKN02055267 AB062937 KB632220 ERL90185.1 KN124115 KFO22166.1 APGK01018645 APGK01018646 KB740081 ENN81628.1 KB361983 ELV13362.1 JH000401 EGV98266.1 CABD030064957 ADFV01118439 AQIB01021204 AJFE02025043 AHZZ02009288 ABGA01345193 ABGA01345194 ABGA01349723 NDHI03003383 PNJ71496.1 AQIA01025556 CM001290 EHH57506.1 JSUE03015555 JSUE03015556 JU335113 JV043981 CM001267 AFE78866.1 AFI34052.1 EHH24264.1 AACZ04040353 NBAG03000341 PNI38298.1 GAMT01010251 GAMS01009146 GAMR01002007 GAMP01002235 JAB01610.1 JAB13990.1 JAB31925.1 JAB50520.1 AB076889 AK122986 AL353619 CH471089 BC008065 JH173079 GEBF01005758 EHB15736.1 JAN97874.1 AAGW02058260 DS235092 EEB11794.1 AABR07027065 AK041534 AK136096 AK136101 BAE20592.1 BAE22817.1 BC086799 AEMK02000095 DQIR01277762 HDC33240.1 NTJE010021989 MBV95545.1 AAPE02046211 KE164486 EPQ18215.1 CH916374 EDV91388.1 AGTP01106592 CR860413 AEYP01095441 KE540090 EPY72411.1 AAQR03117180 AAEX03000636 ACTA01075239 GL192650 EFB16599.1 KB099365 ELK38158.1 AANG04003916 GFTR01003306 JAW13120.1 DQ915499 ABJ09404.1 APCN01006053 AAAB01008810 EAA04824.4
KPI97851.1 KQ460883 KPJ11433.1 AGBW02010414 OWR48556.1 JTDY01003999 KOB68731.1 CVRI01000070 CRL07207.1 KQ971372 EFA10311.1 CH902617 EDV42154.1 CM000070 EAL27721.2 KRT00249.1 KRT00250.1 GEZM01103095 GEZM01103093 JAV51564.1 AE014297 BT149876 AAF54453.1 AGC12536.1 ALI30550.1 CM000160 EDW96731.1 CH954181 EDV49705.1 GAMC01008633 GAMC01008632 GAMC01008631 GAMC01008630 JAB97923.1 OUUW01000013 SPP87801.1 CH964232 EDW81261.1 GAKP01022780 JAC36172.1 CH940650 EDW68075.1 AAKN02055267 AB062937 KB632220 ERL90185.1 KN124115 KFO22166.1 APGK01018645 APGK01018646 KB740081 ENN81628.1 KB361983 ELV13362.1 JH000401 EGV98266.1 CABD030064957 ADFV01118439 AQIB01021204 AJFE02025043 AHZZ02009288 ABGA01345193 ABGA01345194 ABGA01349723 NDHI03003383 PNJ71496.1 AQIA01025556 CM001290 EHH57506.1 JSUE03015555 JSUE03015556 JU335113 JV043981 CM001267 AFE78866.1 AFI34052.1 EHH24264.1 AACZ04040353 NBAG03000341 PNI38298.1 GAMT01010251 GAMS01009146 GAMR01002007 GAMP01002235 JAB01610.1 JAB13990.1 JAB31925.1 JAB50520.1 AB076889 AK122986 AL353619 CH471089 BC008065 JH173079 GEBF01005758 EHB15736.1 JAN97874.1 AAGW02058260 DS235092 EEB11794.1 AABR07027065 AK041534 AK136096 AK136101 BAE20592.1 BAE22817.1 BC086799 AEMK02000095 DQIR01277762 HDC33240.1 NTJE010021989 MBV95545.1 AAPE02046211 KE164486 EPQ18215.1 CH916374 EDV91388.1 AGTP01106592 CR860413 AEYP01095441 KE540090 EPY72411.1 AAQR03117180 AAEX03000636 ACTA01075239 GL192650 EFB16599.1 KB099365 ELK38158.1 AANG04003916 GFTR01003306 JAW13120.1 DQ915499 ABJ09404.1 APCN01006053 AAAB01008810 EAA04824.4
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000192223
+ More
UP000183832 UP000007266 UP000007801 UP000001819 UP000192221 UP000000803 UP000002282 UP000008711 UP000268350 UP000007798 UP000008792 UP000189704 UP000005447 UP000233100 UP000030742 UP000028990 UP000019118 UP000248480 UP000011518 UP000001075 UP000233120 UP000233180 UP000001519 UP000001073 UP000233140 UP000029965 UP000240080 UP000028761 UP000233020 UP000233200 UP000233060 UP000233080 UP000233040 UP000233220 UP000001595 UP000009130 UP000006718 UP000002277 UP000008225 UP000005640 UP000006813 UP000081671 UP000001811 UP000009046 UP000002494 UP000189706 UP000000589 UP000002281 UP000008227 UP000265300 UP000248484 UP000261681 UP000001074 UP000007646 UP000001070 UP000005215 UP000079721 UP000252040 UP000245320 UP000248483 UP000245341 UP000248482 UP000245340 UP000000715 UP000248481 UP000286641 UP000005225 UP000233160 UP000261680 UP000291021 UP000002254 UP000286640 UP000286642 UP000008912 UP000011712 UP000075901 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062
UP000183832 UP000007266 UP000007801 UP000001819 UP000192221 UP000000803 UP000002282 UP000008711 UP000268350 UP000007798 UP000008792 UP000189704 UP000005447 UP000233100 UP000030742 UP000028990 UP000019118 UP000248480 UP000011518 UP000001075 UP000233120 UP000233180 UP000001519 UP000001073 UP000233140 UP000029965 UP000240080 UP000028761 UP000233020 UP000233200 UP000233060 UP000233080 UP000233040 UP000233220 UP000001595 UP000009130 UP000006718 UP000002277 UP000008225 UP000005640 UP000006813 UP000081671 UP000001811 UP000009046 UP000002494 UP000189706 UP000000589 UP000002281 UP000008227 UP000265300 UP000248484 UP000261681 UP000001074 UP000007646 UP000001070 UP000005215 UP000079721 UP000252040 UP000245320 UP000248483 UP000245341 UP000248482 UP000245340 UP000000715 UP000248481 UP000286641 UP000005225 UP000233160 UP000261680 UP000291021 UP000002254 UP000286640 UP000286642 UP000008912 UP000011712 UP000075901 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9ISS6
A0A2W1BJA6
A0A2H1V241
A0A194PWW5
A0A194R0W0
A0A212F489
+ More
A0A0L7L0A3 A0A1W4WM32 A0A1J1J484 D6X324 B3M1C7 Q299I2 A0A1Y1JQS7 A0A1W4W0V2 Q9VH66 B4PVF8 B3P4Q2 W8BLM8 A0A3B0KRD2 B4N810 A0A034V216 B4LYX2 A0A1U7UQZ4 A0A286XUM9 Q95KD9 U4UJK8 A0A091DHC1 N6TUB6 A0A2Y9E698 L8YAR4 G3HIA6 A0A2K6AND2 A0A2K6JVH6 A0A2I2Y9Y4 A0A2I3HQU6 A0A2K5XXR9 A0A0D9SB09 A0A2R8ZHX2 A0A096NE98 A0A2K5DZN0 A0A2K6NIW5 A0A2K5L062 A0A2K5H7K0 A0A2K5PA64 A0A2K6S7X6 H2PSM8 G7PSP0 F7EK27 A0A2I3SKK7 F7FTA1 Q96HU8 G5C2H5 A0A1S3G0J3 G1SCX2 E0VEI8 D3ZHX3 Q3UWU7 A0A1U8BF78 Q5PR73 F7DZR6 A0A287AKH0 A0A340WB13 A0A2Y9FDB2 A0A2F0AXQ5 A0A384B2G5 G1PCU9 S7NNM2 G3TH62 B4JV68 I3MX94 Q5R6S2 A0A1S3AT03 A0A341ACT5 A0A2U4BF66 A0A2Y9M5Q2 A0A2U3XPN3 A0A2Y9K525 A0A2U3WLR0 M3Z710 S9XB92 A0A2Y9HE79 A0A3Q7QMT5 H0XIA3 A0A2K6GK24 A0A384C2L9 E2RK40 A0A3Q7RCR3 A0A3Q7Y3A8 D2HB69 L5MK87 M3WE12 A0A224XX81 A0A182T4X9 Q007S9 A0A182WS04 A0A182UYI2 A0A182KVY0 A0A182TWS6 A0A182ICW6 Q7QI45
A0A0L7L0A3 A0A1W4WM32 A0A1J1J484 D6X324 B3M1C7 Q299I2 A0A1Y1JQS7 A0A1W4W0V2 Q9VH66 B4PVF8 B3P4Q2 W8BLM8 A0A3B0KRD2 B4N810 A0A034V216 B4LYX2 A0A1U7UQZ4 A0A286XUM9 Q95KD9 U4UJK8 A0A091DHC1 N6TUB6 A0A2Y9E698 L8YAR4 G3HIA6 A0A2K6AND2 A0A2K6JVH6 A0A2I2Y9Y4 A0A2I3HQU6 A0A2K5XXR9 A0A0D9SB09 A0A2R8ZHX2 A0A096NE98 A0A2K5DZN0 A0A2K6NIW5 A0A2K5L062 A0A2K5H7K0 A0A2K5PA64 A0A2K6S7X6 H2PSM8 G7PSP0 F7EK27 A0A2I3SKK7 F7FTA1 Q96HU8 G5C2H5 A0A1S3G0J3 G1SCX2 E0VEI8 D3ZHX3 Q3UWU7 A0A1U8BF78 Q5PR73 F7DZR6 A0A287AKH0 A0A340WB13 A0A2Y9FDB2 A0A2F0AXQ5 A0A384B2G5 G1PCU9 S7NNM2 G3TH62 B4JV68 I3MX94 Q5R6S2 A0A1S3AT03 A0A341ACT5 A0A2U4BF66 A0A2Y9M5Q2 A0A2U3XPN3 A0A2Y9K525 A0A2U3WLR0 M3Z710 S9XB92 A0A2Y9HE79 A0A3Q7QMT5 H0XIA3 A0A2K6GK24 A0A384C2L9 E2RK40 A0A3Q7RCR3 A0A3Q7Y3A8 D2HB69 L5MK87 M3WE12 A0A224XX81 A0A182T4X9 Q007S9 A0A182WS04 A0A182UYI2 A0A182KVY0 A0A182TWS6 A0A182ICW6 Q7QI45
PDB
2ERX
E-value=8.82546e-73,
Score=692
Ontologies
GO
PANTHER
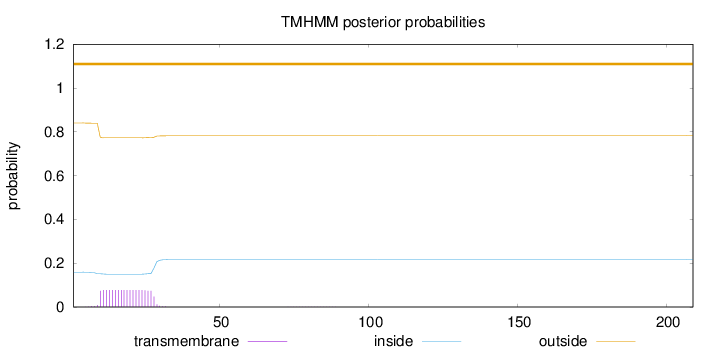
Topology
Subcellular location
Cell membrane
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.47706
Exp number, first 60 AAs:
1.47284
Total prob of N-in:
0.15920
outside
1 - 209
Population Genetic Test Statistics
Pi
118.48386
Theta
193.019474
Tajima's D
-1.418357
CLR
1336.642658
CSRT
0.072246387680616
Interpretation
Uncertain
