Gene
KWMTBOMO13076
Annotation
hypothetical_protein_RR48_15076_[Papilio_machaon]
Full name
GPN-loop GTPase 2
+ More
Proteasome assembly chaperone 3
Proteasome assembly chaperone 3
Location in the cell
Cytoplasmic Reliability : 2.032 Nuclear Reliability : 2.153
Sequence
CDS
ATGGATCTCCCAGATATTATGAAAAATTTGAAAATACAAAATTCTGAAGCCCCAAATAATTTATTTAAATCAGTGAGTGCTGAAATAGATGATGTCACTACTGATATAGTGATAGCAGAATGTAGTGATAAAGTGTTTTTAGTTGTATCTCAGTATCAAAAAATGGGAAGTATGCTAATGGTTGTCAGAGATCGAATCAACGGTCCTCATGGCATTGAAGATGTGTATTCAACAAAAGTTGTTTTTGGTGATTCAGGGGAAGAGCATCAAGCAGCAGCAAGATTCTTAGCTGAAACAGTAGATATATTATCAAAACCACTTTGCATTTTCATCAACTTAAGGTCATATGACATTGAAACACTGAAAGCTTGTAGAGATATCATTTTGGACTTCAAAAAAGAAGATACAGAATGTCAGCAATGA
Protein
MDLPDIMKNLKIQNSEAPNNLFKSVSAEIDDVTTDIVIAECSDKVFLVVSQYQKMGSMLMVVRDRINGPHGIEDVYSTKVVFGDSGEEHQAAARFLAETVDILSKPLCIFINLRSYDIETLKACRDIILDFKKEDTECQQ
Summary
Description
Small GTPase required for proper localization of RNA polymerase II and III (RNAPII and RNAPIII). May act at an RNAP assembly step prior to nuclear import.
Chaperone protein which promotes assembly of the 20S proteasome. May cooperate with psmg1-psmg2 heterodimers to orchestrate the correct assembly of proteasomes (By similarity).
Chaperone protein which promotes assembly of the 20S proteasome. May cooperate with psmg1-psmg2 heterodimers to orchestrate the correct assembly of proteasomes (By similarity).
Subunit
Binds to RNA polymerase II (RNAPII).
Similarity
Belongs to the GPN-loop GTPase family.
Belongs to the PSMG3 family.
Belongs to the PSMG3 family.
Keywords
Chaperone
Complete proteome
Reference proteome
Feature
chain Proteasome assembly chaperone 3
Uniprot
A0A194R156
A0A194PWZ8
A0A232EZ49
A0A1Y1NHB4
A0A1B6L0I1
A0A0N0U5B6
+ More
A0A0K8RH61 B7PJR2 A0A131YA39 A0A310S3X4 A0A293LP11 A0A088AH44 A0A1S3KEC3 A0A1S3HXR7 A0A1Z5L8J6 E9FTY8 A0A0P4Y224 A0A154PDL1 A0A0P6I3L1 A0A1E1WZN8 C3YZA5 H3AN86 A0A1S4EJT6 A0A060WX84 B5X7K7 A0A3P9AFJ1 A0A2G8JYH9 B5X5K2 A0A315V3M4 A0A2I4AIP8 Q6DGH7 A0A2C9LY96 A0A3B5A7Q1 A0A060WHM5 A0A0A0KUG9 A9NRF5 R7QMC1 A0A3P8X0A7 G3PM10 A0A3Q3B8L1 A0A096M830 A0A3B3VKQ5 A0A3B3WFG8 A0A1S3AUA3 M4A4F0 A0A3B5LM36 A0A084WBG0 A0A3B3SET4 A0A1X0RM75 A0A367J727 C3KK99 A0A124SHC7 A0A3P8THF7 A0A3Q1B5H2 W9SNR9 A0A182QW99 A0A3Q1GUP5 A0A3Q0R6P7 A0A182MXX7 A0A3Q1HCD0 A0A3Q3GJ18 G7J4Z4 W9RBM2 A0A0E9XHI8 A0A3Q4BXE8 A0A1J3CMK7 A7SP74 A0A3B5K109 A0A2N9J0B1 A0A182SZV2 A0A3Q0RAQ9 A0A182RFB9 A0A182JYW4 A0A1A7Z3G8 A0A178U9J1 Q9LER5 A0A182YDB9 A0A3B4V3L7 A0A3Q3KMR2 A0A2K3MVI8 A0A3Q3K9Y4 D7M6M2 R0HBD2
A0A0K8RH61 B7PJR2 A0A131YA39 A0A310S3X4 A0A293LP11 A0A088AH44 A0A1S3KEC3 A0A1S3HXR7 A0A1Z5L8J6 E9FTY8 A0A0P4Y224 A0A154PDL1 A0A0P6I3L1 A0A1E1WZN8 C3YZA5 H3AN86 A0A1S4EJT6 A0A060WX84 B5X7K7 A0A3P9AFJ1 A0A2G8JYH9 B5X5K2 A0A315V3M4 A0A2I4AIP8 Q6DGH7 A0A2C9LY96 A0A3B5A7Q1 A0A060WHM5 A0A0A0KUG9 A9NRF5 R7QMC1 A0A3P8X0A7 G3PM10 A0A3Q3B8L1 A0A096M830 A0A3B3VKQ5 A0A3B3WFG8 A0A1S3AUA3 M4A4F0 A0A3B5LM36 A0A084WBG0 A0A3B3SET4 A0A1X0RM75 A0A367J727 C3KK99 A0A124SHC7 A0A3P8THF7 A0A3Q1B5H2 W9SNR9 A0A182QW99 A0A3Q1GUP5 A0A3Q0R6P7 A0A182MXX7 A0A3Q1HCD0 A0A3Q3GJ18 G7J4Z4 W9RBM2 A0A0E9XHI8 A0A3Q4BXE8 A0A1J3CMK7 A7SP74 A0A3B5K109 A0A2N9J0B1 A0A182SZV2 A0A3Q0RAQ9 A0A182RFB9 A0A182JYW4 A0A1A7Z3G8 A0A178U9J1 Q9LER5 A0A182YDB9 A0A3B4V3L7 A0A3Q3KMR2 A0A2K3MVI8 A0A3Q3K9Y4 D7M6M2 R0HBD2
Pubmed
26354079
28648823
28004739
28528879
21292972
28503490
+ More
18563158 9215903 24755649 20433749 25069045 29023486 29703783 23594743 15562597 19881527 19495411 20565788 22047402 18854048 23503846 24487278 22753475 23542700 24438588 29240929 27956601 29674435 26786968 22089132 24767513 30397259 25613341 17615350 21551351 22223895 27354520 11130714 12093376 27862469 25244985 24500806 28382043 21478890 23749190
18563158 9215903 24755649 20433749 25069045 29023486 29703783 23594743 15562597 19881527 19495411 20565788 22047402 18854048 23503846 24487278 22753475 23542700 24438588 29240929 27956601 29674435 26786968 22089132 24767513 30397259 25613341 17615350 21551351 22223895 27354520 11130714 12093376 27862469 25244985 24500806 28382043 21478890 23749190
EMBL
KQ460883
KPJ11437.1
KQ459586
KPI97847.1
NNAY01001524
OXU23694.1
+ More
GEZM01005619 JAV96035.1 GEBQ01022765 JAT17212.1 KQ435794 KOX73978.1 GADI01003338 JAA70470.1 ABJB010201090 DS727794 EEC06834.1 GEFM01000486 JAP75310.1 KQ797310 OAD46961.1 GFWV01010508 MAA35237.1 GFJQ02003345 JAW03625.1 GL732524 EFX89448.1 GDIP01243022 JAI80379.1 KQ434879 KZC09943.1 GDIQ01026504 JAN68233.1 GFAC01006746 JAT92442.1 GG666566 EEN54176.1 AFYH01097916 FR904779 CDQ71617.1 BT047026 ACI66827.1 MRZV01001083 PIK40798.1 BT046321 BT125413 ACI66122.1 ADM16154.1 NHOQ01002357 PWA17712.1 BX908797 BC076368 BC165128 AAH76368.1 AAI65128.1 FR904459 CDQ64070.1 CM002926 KGN51386.1 EF083883 EF086424 ABK23216.1 HG002061 CDF39657.1 AYCK01008293 ATLV01022351 KE525331 KFB47554.1 KV921564 ORE13135.1 PJQL01002018 RCH85772.1 BT083364 ACQ59071.1 LEKV01001069 KVI09163.1 KE619967 EXC36383.1 AXCN02002113 CM001219 PSQE01000003 AES72307.1 RHN69456.1 KE343942 EXB48383.1 GBXM01007414 JAI01164.1 GEVI01023151 JAU09169.1 DS469729 OIVN01006293 SPD29940.1 HADW01004563 HADX01014766 SBP36998.1 LUHQ01000005 OAO89807.1 AY084903 BT005169 CP002688 AK117635 AL391149 AAM61466.1 AAO50702.1 AED92065.1 BAC42291.1 CAC01870.1 ASHM01012850 PNX94815.1 GL348718 EFH47886.1 KB870810 EOA22335.1
GEZM01005619 JAV96035.1 GEBQ01022765 JAT17212.1 KQ435794 KOX73978.1 GADI01003338 JAA70470.1 ABJB010201090 DS727794 EEC06834.1 GEFM01000486 JAP75310.1 KQ797310 OAD46961.1 GFWV01010508 MAA35237.1 GFJQ02003345 JAW03625.1 GL732524 EFX89448.1 GDIP01243022 JAI80379.1 KQ434879 KZC09943.1 GDIQ01026504 JAN68233.1 GFAC01006746 JAT92442.1 GG666566 EEN54176.1 AFYH01097916 FR904779 CDQ71617.1 BT047026 ACI66827.1 MRZV01001083 PIK40798.1 BT046321 BT125413 ACI66122.1 ADM16154.1 NHOQ01002357 PWA17712.1 BX908797 BC076368 BC165128 AAH76368.1 AAI65128.1 FR904459 CDQ64070.1 CM002926 KGN51386.1 EF083883 EF086424 ABK23216.1 HG002061 CDF39657.1 AYCK01008293 ATLV01022351 KE525331 KFB47554.1 KV921564 ORE13135.1 PJQL01002018 RCH85772.1 BT083364 ACQ59071.1 LEKV01001069 KVI09163.1 KE619967 EXC36383.1 AXCN02002113 CM001219 PSQE01000003 AES72307.1 RHN69456.1 KE343942 EXB48383.1 GBXM01007414 JAI01164.1 GEVI01023151 JAU09169.1 DS469729 OIVN01006293 SPD29940.1 HADW01004563 HADX01014766 SBP36998.1 LUHQ01000005 OAO89807.1 AY084903 BT005169 CP002688 AK117635 AL391149 AAM61466.1 AAO50702.1 AED92065.1 BAC42291.1 CAC01870.1 ASHM01012850 PNX94815.1 GL348718 EFH47886.1 KB870810 EOA22335.1
Proteomes
UP000053240
UP000053268
UP000215335
UP000053105
UP000001555
UP000005203
+ More
UP000085678 UP000000305 UP000076502 UP000001554 UP000008672 UP000079169 UP000193380 UP000087266 UP000265140 UP000230750 UP000192220 UP000000437 UP000076420 UP000261400 UP000029981 UP000012073 UP000265120 UP000007635 UP000264800 UP000028760 UP000261500 UP000261480 UP000089565 UP000002852 UP000261380 UP000030765 UP000261540 UP000242381 UP000252139 UP000243975 UP000265080 UP000257160 UP000030645 UP000075886 UP000257200 UP000261340 UP000075884 UP000265040 UP000261660 UP000002051 UP000265566 UP000261620 UP000001593 UP000005226 UP000075901 UP000075900 UP000075881 UP000078284 UP000006548 UP000076408 UP000261420 UP000261640 UP000236291 UP000261600 UP000008694 UP000029121
UP000085678 UP000000305 UP000076502 UP000001554 UP000008672 UP000079169 UP000193380 UP000087266 UP000265140 UP000230750 UP000192220 UP000000437 UP000076420 UP000261400 UP000029981 UP000012073 UP000265120 UP000007635 UP000264800 UP000028760 UP000261500 UP000261480 UP000089565 UP000002852 UP000261380 UP000030765 UP000261540 UP000242381 UP000252139 UP000243975 UP000265080 UP000257160 UP000030645 UP000075886 UP000257200 UP000261340 UP000075884 UP000265040 UP000261660 UP000002051 UP000265566 UP000261620 UP000001593 UP000005226 UP000075901 UP000075900 UP000075881 UP000078284 UP000006548 UP000076408 UP000261420 UP000261640 UP000236291 UP000261600 UP000008694 UP000029121
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A194R156
A0A194PWZ8
A0A232EZ49
A0A1Y1NHB4
A0A1B6L0I1
A0A0N0U5B6
+ More
A0A0K8RH61 B7PJR2 A0A131YA39 A0A310S3X4 A0A293LP11 A0A088AH44 A0A1S3KEC3 A0A1S3HXR7 A0A1Z5L8J6 E9FTY8 A0A0P4Y224 A0A154PDL1 A0A0P6I3L1 A0A1E1WZN8 C3YZA5 H3AN86 A0A1S4EJT6 A0A060WX84 B5X7K7 A0A3P9AFJ1 A0A2G8JYH9 B5X5K2 A0A315V3M4 A0A2I4AIP8 Q6DGH7 A0A2C9LY96 A0A3B5A7Q1 A0A060WHM5 A0A0A0KUG9 A9NRF5 R7QMC1 A0A3P8X0A7 G3PM10 A0A3Q3B8L1 A0A096M830 A0A3B3VKQ5 A0A3B3WFG8 A0A1S3AUA3 M4A4F0 A0A3B5LM36 A0A084WBG0 A0A3B3SET4 A0A1X0RM75 A0A367J727 C3KK99 A0A124SHC7 A0A3P8THF7 A0A3Q1B5H2 W9SNR9 A0A182QW99 A0A3Q1GUP5 A0A3Q0R6P7 A0A182MXX7 A0A3Q1HCD0 A0A3Q3GJ18 G7J4Z4 W9RBM2 A0A0E9XHI8 A0A3Q4BXE8 A0A1J3CMK7 A7SP74 A0A3B5K109 A0A2N9J0B1 A0A182SZV2 A0A3Q0RAQ9 A0A182RFB9 A0A182JYW4 A0A1A7Z3G8 A0A178U9J1 Q9LER5 A0A182YDB9 A0A3B4V3L7 A0A3Q3KMR2 A0A2K3MVI8 A0A3Q3K9Y4 D7M6M2 R0HBD2
A0A0K8RH61 B7PJR2 A0A131YA39 A0A310S3X4 A0A293LP11 A0A088AH44 A0A1S3KEC3 A0A1S3HXR7 A0A1Z5L8J6 E9FTY8 A0A0P4Y224 A0A154PDL1 A0A0P6I3L1 A0A1E1WZN8 C3YZA5 H3AN86 A0A1S4EJT6 A0A060WX84 B5X7K7 A0A3P9AFJ1 A0A2G8JYH9 B5X5K2 A0A315V3M4 A0A2I4AIP8 Q6DGH7 A0A2C9LY96 A0A3B5A7Q1 A0A060WHM5 A0A0A0KUG9 A9NRF5 R7QMC1 A0A3P8X0A7 G3PM10 A0A3Q3B8L1 A0A096M830 A0A3B3VKQ5 A0A3B3WFG8 A0A1S3AUA3 M4A4F0 A0A3B5LM36 A0A084WBG0 A0A3B3SET4 A0A1X0RM75 A0A367J727 C3KK99 A0A124SHC7 A0A3P8THF7 A0A3Q1B5H2 W9SNR9 A0A182QW99 A0A3Q1GUP5 A0A3Q0R6P7 A0A182MXX7 A0A3Q1HCD0 A0A3Q3GJ18 G7J4Z4 W9RBM2 A0A0E9XHI8 A0A3Q4BXE8 A0A1J3CMK7 A7SP74 A0A3B5K109 A0A2N9J0B1 A0A182SZV2 A0A3Q0RAQ9 A0A182RFB9 A0A182JYW4 A0A1A7Z3G8 A0A178U9J1 Q9LER5 A0A182YDB9 A0A3B4V3L7 A0A3Q3KMR2 A0A2K3MVI8 A0A3Q3K9Y4 D7M6M2 R0HBD2
Ontologies
PANTHER
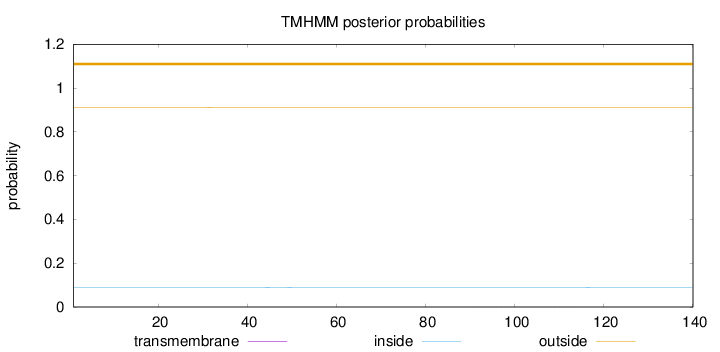
Topology
Length:
140
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00587
Exp number, first 60 AAs:
0.00169
Total prob of N-in:
0.08849
outside
1 - 140
Population Genetic Test Statistics
Pi
320.51899
Theta
215.297832
Tajima's D
1.268126
CLR
0
CSRT
0.728863556822159
Interpretation
Uncertain
